Abstract
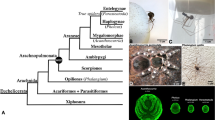
Despite application of genome-scale datasets, the phylogenetic placement of scorpions within arachnids remains contentious between two different phylogenetic data classes. Paleontologists continue to recover scorpions in a basally branching position, partly owing to their morphological similarity to extinct marine orders like Eurypterida (sea scorpions). Phylogenomic datasets consistently recover scorpions in a derived position, as the sister group of Tetrapulmonata (a clade of arachnids that includes spiders). To adjudicate between these hypotheses using a rare genomic change (RGC), we leveraged the recent discovery of ancient paralogy in spiders and scorpions to assess phylogenetic placement. We identified homologs of four transcription factors required for appendage patterning (dachshund, homothorax, extradenticle, and optomotor blind) in arthropods that are known to be duplicated in spiders. Using genomic resources for a spider, a scorpion, and a harvestman, we conducted gene tree analyses and assayed expression patterns of scorpion gene duplicates. Here we show that scorpions, like spiders, retain two copies of all four transcription factors, whereas arachnid orders like mites and harvestmen bear a single copy. A survey of embryonic expression patterns of the scorpion paralogs closely matches those of their spider counterparts, with one paralog consistently retaining the putatively ancestral pattern found in the harvestman, as well as the mite, and/or other outgroups. These data comprise a rare genomic change in chelicerate phylogeny supporting the inference of a distal placement of scorpions. Beyond demonstrating the diagnostic power of developmental genetic data as a phylogenetic data class, a derived placement of scorpions within the arachnids, together with an array of stem-group Paleozoic scorpions that occupied marine habitats, effectively rules out a scenario of a single colonization of terrestrial habitat within Chelicerata, even in tree topologies contrived to recover the monophyly of Arachnida.








Similar content being viewed by others
References
Aria C, Caron J-B (2019) A middle Cambrian arthropod with chelicerae and proto-book gills. Nature 465:215–214. https://doi.org/10.1038/s41586-019-1525-4
Ballesteros JA, Sharma PP (2019) A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error
Ballesteros JA, Santibáñez-López CE, Kováč L, Gavish-Regev E, Sharma PP (2019) Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi. Proc R Soc Lond B. https://doi.org/10.1098/rspb.2019.2426
Barnett AA, Thomas RH (2013) The expression of limb gap genes in the mite Archegozetes longisetosusreveals differential patterning mechanisms in chelicerates. Evol Dev 15:280–292. https://doi.org/10.1111/ede.12038
Bergsten J (2005) A review of long-branch attraction. Cladistics 21:163–193. https://doi.org/10.1111/j.1096-0031.2005.00059.x
Bicknell RDC, Lustri L, Brougham T (2019) Revision of “Bellinurus” carteri (Chelicerata: Xiphosura) from the Late Devonian of Pennsylvania, USA. Comptes Rendus Palevol. https://doi.org/10.1016/j.crpv.2019.08.002
Bieler R, Mikkelsen PM, Collins TM, Glover EA, González VL, Graf DL, Harper EM, Healy J, Kawauchi GY, Sharma PP, Staubach S, Strong EE, Taylor JD, Temkin I, Zardus JD, Clark S, Guzmán A, McIntyre E, Sharp P, Giribet G (2014) Investigating the bivalve Tree of Life—an exemplar-based approach combining molecular and novel morphological characters. Invertebr Syst 28:32–115. https://doi.org/10.1071/IS13010
Briggs DEG, Siveter DJ, Siveter DJ, Sutton MD, Garwood RJ, Legg D (2012) Silurian horseshoe crab illuminates the evolution of arthropod limbs. Proc Natl Acad Sci U S A 109:15702–15705. https://doi.org/10.1073/pnas.1205875109
Burbrink FT, Grazziotin FG, Pyron RA, Cundall D, Donnellan S, Irish F, Keogh JS, Kraus F, Murphy RW, Noonan B, Raxworthy CJ, Ruane S, Lemmon AR, Lemmon EM, Zaher H (2019) Interrogating genomic-scale data for Squamata (lizards, snakes, and Amphisbaenians) shows no support for key traditional morphological relationships. Syst Biol. https://doi.org/10.1093/sysbio/syz062
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552
Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:e314. https://doi.org/10.1371/journal.pbio.0030314
Di Z, Edgecombe GD, Sharma PP (2018) Homeosis in a scorpion supports a telopodal origin of pectines and components of the book lungs. BMC Evol Biol 18:73. https://doi.org/10.1186/s12862-018-1188-z
Dunlop J (1998) The origins of tetrapulmonate book lungs and their significance for chelicerate phylogeny. Proceedings of the 17th European Colloquium of Arachnology 9–16
Dunlop JA (2010) Geological history and phylogeny of Chelicerata. Arthropod Structure and Development 39:124–142. https://doi.org/10.1016/j.asd.2010.01.003
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleis Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Fröbius AC, Funch P (2017) Rotiferan Hox genes give new insights into the evolution of metazoan bodyplans. Nat Commun 8:9. https://doi.org/10.1038/s41467-017-00020-w
Garwood RJ, Dunlop J (2014) Three-dimensional reconstruction and the phylogeny of extinct chelicerate orders. PeerJ 2:e641. https://doi.org/10.7717/peerj.641/supp-4
Garwood RJ, Dunlop JA, Selden PA, Spencer AR, Atwood RC, Vo NT, Drakopoulos M (2016) Almost a spider: a 305-million-year-old fossil arachnid and spider origins. Proc R Soc B Biol Sci 283:20160125–20160128. https://doi.org/10.1098/rspb.2016.0125
Goloboff PA, Catalano SA (2016) TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32:221–238. https://doi.org/10.1111/cla.12160
Hazkani-Covo E (2009) Mitochondrial insertions into primate nuclear genomes suggest the use of numts as a tool for phylogeny. Mol Biol Evol 26:2175–2179. https://doi.org/10.1093/molbev/msp131
Howard RJ, Edgecombe GD, Legg DA et al (2019) Exploring the evolution and terrestrialization of scorpions (Arachnida: Scorpiones) with rocks and clocks:1–16. https://doi.org/10.1007/s13127-019-00390-7
Huang D, Hormiga G, Cai C, Su Y, Yin Z, Xia F, Giribet G (2018) Origin of spiders and their spinning organs illuminated by mid-Cretaceous amber fossils. Nat Ecol Evol 2:623–627. https://doi.org/10.1038/s41559-018-0475-9
Janssen R, Feitosa NM, Damen WGM, Prpic N-M (2008) The T-box genes H15 and optomotor-blind in the spiders Cupiennius salei, Tegenaria atrica and Achaearanea tepidariorum and the dorsoventral axis of arthropod appendages. Evol Dev 10:143–154. https://doi.org/10.1111/j.1525-142X.2008.00222.x
Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, Chan TF, Kwan HS, Holland PW, Chu KH, Hui JH (2016) Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity 116:190–199. https://doi.org/10.1038/hdy.2015.89
Klußmann-Fricke BJ, Wirkner CS (2016) Comparative morphology of the hemolymph vascular system in Uropygi and Amblypygi (Arachnida): complex correspondences support Arachnopulmonata. J Morphol:1–20. https://doi.org/10.1002/jmor.20559
Klußmann-Fricke BJ, Pomrehn SW, Wirkner CS (2014) A wonderful network unraveled-detailed description of capillaries in the prosomal ganglion of scorpions. Front Zool 11:28. https://doi.org/10.1186/1742-9994-11-28
Lamsdell JC (2016) Horseshoe crab phylogeny and independent colonizations of fresh water: ecological invasion as a driver for morphological innovation. Palaeontology 59:181–194. https://doi.org/10.1111/pala.12220
Lamsdell JC, Briggs DEG, Liu HP, Witzke BJ, McKay RM (2015) A new Ordovician arthropod from the Winneshiek Lagerstätte of Iowa (USA) reveals the ground plan of eurypterids and chasmataspidids. Sci Nat- Heidelberg 102:63. https://doi.org/10.1007/s00114-015-1312-5
Lartillot N, Philippe H (2004) A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21(6):1095–1109. https://doi.org/10.1093/molbev/msh112
Lartillot N, Brinkmann H, Philippe H (2007) Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7:S4. https://doi.org/10.1186/1471-2148-7-S1-S4
Legg DA, Sutton MD, Edgecombe GD (2013) Arthropod fossil data increase congruence of morphological and molecular phylogenies. Nat Commun 4:2485. https://doi.org/10.1038/ncomms3485
Lehmann T, Melzer RR (2019) The visual system of Thelyphonida (whip scorpions): support for Arachnopulmonata. Arthropod Struct Dev 51:23–31. https://doi.org/10.1016/j.asd.2019.06.002
Leite DJ, Ninova M, Hilbrant M, Arif S, Griffiths-Jones S, Ronshaugen M, McGregor A (2016) Pervasive microRNA duplication in Chelicerates: insights from the embryonic microRNA repertoire of the spider Parasteatoda tepidariorum. Genome Biol Evol 8:2133–2144. https://doi.org/10.1093/gbe/evw143
Leite DJ, Baudouin-Gonzalez L, Iwasaki-Yokozawa S et al (2018) Homeobox gene duplication and divergence in arachnids. Mol Biol Evol 35:2240–2253. https://doi.org/10.1093/molbev/msy125
Liao YY, Xu PW, Kwan KY, Ma ZY, Fang HY, Xu JY, Wang PL, Yang SY, Xie SB, Xu SQ, Qian D, Li WF, Bai LR, Zhou DJ, Zhang YQ, Lei J, Liu K, Li F, Li J, Zhu P, Wang YJ, Wu HP, Xu YH, Huang H, Zhang C, Liu JX, Han JF (2019) Draft genomic and transcriptome resources for marine chelicerate Tachypleus tridentatus. Nat Publ Group 6:1–10. https://doi.org/10.1038/sdata.2019.29
Lozano-Fernandez J, Tanner AR, Giacomelli M, Carton R, Vinther J, Edgecombe GD, Pisani D (2019) Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat Commun 10:1–8. https://doi.org/10.1038/s41467-019-10244-7
Meusemann K, Reumont Von BM, Simon S et al (2010) A Phylogenomic approach to resolve the arthropod Tree of Life. Mol Biol Evol 27:2451–2464. https://doi.org/10.1093/molbev/msq130
Monod L, Cauwet L, González-Santillan E, Huber S (2017) The male sexual apparatus in the order Scorpiones (Arachnida): a comparative study of functional morphology as a tool to define hypotheses of homology. Front Zool 14:51. https://doi.org/10.1186/s12983-017-0231-z
Pechmann M, Prpic N-M (2009) Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae). Dev Genes Evol 219:189–198. https://doi.org/10.1007/s00427-009-0279-7
Poschmann M, Dunlop JA, Kamenz C, Scholtz G (2008) The Lower Devonian scorpion Waeringoscorpio and the respiratory nature of its filamentous structures, with the description of a new species from the Westerwald area, Germany. Paläontol Z 82(4):418–436. https://doi.org/10.1007/BF03184431
Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW (2010) Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463:1079–1083. https://doi.org/10.1038/nature08742
Rokas A, Carroll SB (2006) Bushes in the Tree of Life. PLoS Biol 4:1899–1904. https://doi.org/10.1371/journal.pbio.0040352
Rokas A, Holland P (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Ronco M, Uda T, Mito T, Minelli A, Noji S, Klingler M (2008) Antenna and all gnathal appendages are similarly transformed by homothorax knock-down in the cricket Gryllus bimaculatus. Dev Biol 313:80–92. https://doi.org/10.1016/j.ydbio.2007.09.059
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Sucha MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Salichos L, Rokas A (2014) Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497:327–331. https://doi.org/10.1038/nature12130
Santibáñez-López CE, Krievel R, Sharma PP (2017) eadem figura manet: measuring morphological convergence in diplocentrid scorpions (Arachnida: Scorpiones: Diplocentridae) under a multilocus phylogenetic framework. Invertebr Syst 31:233–248. https://doi.org/10.1071/IS16078
Santibáñez-López CE, González-Santillán E, Monod L, Sharma PP (2019) Phylogenomics facilitates stable scorpion systematics: reassessing the relationships of Vaejovidae and a new higher-level classification of Scorpiones (Arachnida). Mol Phylogenet Evol 135:22–30. https://doi.org/10.1016/j.ympev.2019.02.021
Scholtz G, Kamenz C (2006) The book lungs of Scorpiones and Tetrapulmonata (Chelicerata, Arachnida): evidence for homology and a single terrestrialisation event of a common arachnid ancestor. 109:2–13. https://doi.org/10.1016/j.zool.2005.06.003
Schwager EE, Schoppmeier M, Pechmann M, Damen WG (2007) Duplicated Hox genes in the spider Cupiennius salei. Front Zool 4:10. https://doi.org/10.1186/1742-9994-4-10
Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, Buffry AD, Chao H, Dinh H, Doddapaneni H, Dugan S, Eibner C, Extavour CG, Funch P, Garb J, Gonzalez LB, Gonzalez VL, Griffiths-Jones S, Han Y, Hayashi C, Hilbrant M, Hughes DST, Janssen R, Lee SL, Maeso I, Murali SC, Muzny DM, Nunes da Fonseca R, Paese CLB, Qu J, Ronshaugen M, Schomburg C, Schönauer A, Stollewerk A, Torres-Oliva M, Turetzek N, Vanthournout B, Werren JH, Wolff C, Worley KC, Bucher G, Gibbs RA, Coddington J, Oda H, Stanke M, Ayoub NA, Prpic NM, Flot JF, Posnien N, Richards S, McGregor A (2017) The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol 15:62. https://doi.org/10.1186/s12915-017-0399-x
Selden PA, Jeram AJ (1989) Palaeophysiology of terrestrialisation in the Chelicerata. Trans R Soc Edinb Earth Sci 80:303–310
Setton EVW, Sharma PP (2018) Cooption of an appendage-patterning gene cassette in the head segmentation of arachnids. Proc Natl Acad Sci 115:E3491–E3500. https://doi.org/10.1073/pnas.1720193115
Sharma PP (2017) Chelicerates and the conquest of land: a view of arachnid origins through an Evo-Devo spyglass. Integr Comp Biol 57:510–522. https://doi.org/10.1093/icb/icx078
Sharma PP, Schwager EE, Extavour CG, Giribet G (2012a) Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol Dev 14:450–463. https://doi.org/10.1111/j.1525-142X.2012.00565.x
Sharma PP, Schwager EE, Extavour CG, Giribet G (2012b) Evolution of the chelicera: a dachshund domain is retained in the deutocerebral appendage of Opiliones (Arthropoda, Chelicerata). Evol Dev 14:522–533. https://doi.org/10.1111/ede.12005
Sharma PP, Schwager EE, Giribet G, Jockusch EL, Extavour CG (2013) Distal-less and dachshund pattern both plesiomorphic and apomorphic structures in chelicerates: RNA interference in the harvestman Phalangium opilio (Opiliones). Evol Dev 15:228–242. https://doi.org/10.1111/ede.12029
Sharma PP, Kaluziak ST, Perez-Porro AR et al (2014a) Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol 31:2963–2984. https://doi.org/10.1093/molbev/msu235
Sharma PP, Schwager EE, Extavour CG, Wheeler WC (2014b) Hox gene duplications correlate with posterior heteronomy in scorpions. Proc R Soc B Biol Sci 281:20140661–20140661. https://doi.org/10.1016/j.cub.2009.06.061
Sharma PP, Gupta T, Schwager EE, Wheeler WC, Extavour CG (2014c) Subdivision of arthropod cap-n-collar expression domains is restricted to Mandibulata. EvoDevo 5:3. https://doi.org/10.1186/2041-9139-5-3
Sharma PP, Santiago MA, González-Santillán E, Monod L, Wheeler WC (2015a) Evidence of duplicated Hox genes in the most recent common ancestor of extant scorpions. Evol Dev 17:347–355. https://doi.org/10.1111/ede.12166
Sharma PP, Tarazona OA, Lopez DH, Schwager EE, Cohn MJ, Wheeler WC, Extavour CG (2015b) A conserved genetic mechanism specifies deutocerebral appendage identity in insects and arachnids. Proc R Soc B Biol Sci 282:20150698–20150698. https://doi.org/10.1098/rspb.2015.0698
Sharma PP, Fernandez R, Esposito LA et al (2015c) Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal. Proc R Soc B Biol Sci 282:20142953–20142953. https://doi.org/10.1093/molbev/mss208
Sharma PP, Baker CM, Cosgrove JG, Johnson JE, Oberski JT, Raven RJ, Harvey MS, Boyer SL, Giribet G (2018) A revised dated phylogeny of scorpions: phylogenomic support for ancient divergence of the temperate Gondwanan family Bothriuridae. Mol Phylogenet Evol 122:37–45. https://doi.org/10.1016/j.ympev.2018.01.003
Shen X-X, Hittinger CT, Rokas A (2017) Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol 1:1–10. https://doi.org/10.1038/s41559-017-0126
Shultz JW (1990) Evolutionary morphology and phylogeny of Arachnida. Cladistics 6:1–38. https://doi.org/10.1111/j.1096-0031.1990.tb00523.x
Shultz J (2007) A phylogenetic analysis of the arachnid orders based on morphological characters
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Turetzek N, Pechmann M, Schomburg C et al (2015) Neofunctionalization of a duplicate dachshund gene underlies the evolution of a novel leg segment in arachnids. Mol Biol Evol 33:109–121. https://doi.org/10.1093/molbev/msv200
Turetzek N, Khadjeh S, Schomburg C, Prpic N-M (2017) Rapid diversification of homothorax expression patterns after gene duplication in spiders. BMC Evol Biol 17:168. https://doi.org/10.1186/s12862-017-1013-0
Waddington J, Rudkin DM, Dunlop JA (2015) A new mid-Silurian aquatic scorpion—one step closer to land? Biol Lett 11:20140815–20140815. https://doi.org/10.1016/j.palaeo.2008.05.008
Wang B, Dunlop JA, Selden PA, Garwood RJ, Shear WA, Müller P, Lei X (2018) Cretaceous arachnid Chimerarachne yingi gen. Et sp. nov. illuminates spider origins. Nat Ecol Evol 2:614–622. https://doi.org/10.1038/s41559-017-0449-3
Acknowledgments
We are indebted to Angelika Stollewerk and Matthias Pechmann for inviting this contribution. Russell Bicknell kindly provided a morphological data matrix for reanalysis. Melody Albright assisted with field collection of C. sculpturatus. Comments from Emily V.W. Setton, Guilherme Gainett, Jesús A. Ballesteros, and two anonymous reviewers improved a previous draft of the manuscript. Discussions with three colleagues in the paleontological community, Russell Garwood, Jason Dunlop, and Paul Selden, greatly refined some of the discussion on Paleozoic scorpions. James Lamsdell could not be reached for comment.
Funding
This work is based on material supported by National Science Foundation grant IOS-1552610 to PPS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Matthias Pechmann
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Special Issue “Crossroads in Spider Research—evolutionary, ecological, and economic significance”
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nolan, E.D., Santibáñez-López, C.E. & Sharma, P.P. Developmental gene expression as a phylogenetic data class: support for the monophyly of Arachnopulmonata. Dev Genes Evol 230, 137–153 (2020). https://doi.org/10.1007/s00427-019-00644-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-019-00644-6