Abstract
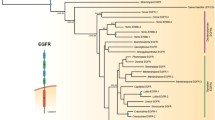
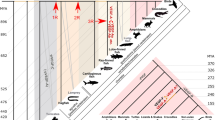
We have analyzed the evolution of fibroblast growth factor receptor (FGFR) tyrosine kinase genes throughout a wide range of animal phyla. No evidence for an FGFR gene was found in Porifera, but we tentatively identified an FGFR gene in the placozoan Trichoplax adhaerens. The gene encodes a protein with three immunoglobulin-like domains, a single-pass transmembrane, and a split tyrosine kinase domain. By superimposing intron positions of 20 FGFR genes from Placozoa, Cnidaria, Protostomia, and Deuterostomia over the respective protein domain structure, we identified ten ancestral introns and three conserved intron groups. Our analysis shows (1) that the position of ancestral introns correlates to the modular structure of FGFRs, (2) that the acidic domain very likely evolved in the last common ancestor of triploblasts, (3) that splicing of IgIII was enabled by a triploblast-specific insertion, and (4) that IgI is subject to substantial loss or duplication particularly in quickly evolving genomes. Moreover, intron positions in the catalytic domain of FGFRs map to the borders of protein subdomains highly conserved in other serine/threonine kinases. Nevertheless, these introns were introduced in metazoan receptor tyrosine kinases exclusively. Our data support the view that protein evolution dating back to the Cambrian explosion took place in such a short time window that only subtle changes in the domain structure are detectable in extant representatives of animal phyla. We propose that the first multidomain FGFR originated in the last common ancestor of Placozoa, Cnidaria, and Bilateria. Additional domains were introduced mainly in the ancestor of triploblasts and in the Ecdysozoa.



Similar content being viewed by others
References
Beermann A, Schröder R (2008) Sites of Fgf signalling and perception during embryogenesis of the beetle Tribolium castaneum. Dev Genes Evol 218:153–167
Bourlat SJ, Nielsen C, Economou AD, Telford MJ (2008) Testing the new animal phylogeny: a phylum level molecular analysis of the animal kingdom. Mol Phylogenet Evol 49:23–31
Bridge D, Cunningham CW, DeSalle R, Buss LW (1995) Class-level relationship in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol 12:679–9
Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D (1997) Of worms and men: an evolutionary perspective on the fibroblast growth factor (FGF) and FGF receptor families. J Mol Evol 44:43–56
D’Aniello S, Iirimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, Bertrand S, Garcia-Fernàndez J (2008) Gene expansion and retention leads to a diverse tyrosine kinase superfamily in Amphioxus. Mol Biol Evol 25:1841–1854
Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298(5601):2157–2167
Freeman RM Jr, Wu M, Cordonnier-Pratt MM, Pratt LH, Gruber CE, Smith M, Lander ES, Stange-Thomann N, Lowe CJ, Gerhart J, Kirschner M (2008) cDNA sequences for transcription factors and signaling proteins of the hemichordate Saccoglossus kowalevskii: efficacy of the expressed sequence tag (EST) approach for evolutionary and developmental studies of a new organism. Biol Bull 14:284–302
Furdui CM, Lew ED, Schlessinger J, Anderson KS (2006) Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell 21:711–717
Gilbert W (1978) Why genes in pieces? Nature 271:501
Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM (1996) heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev 10:3003–3017
Grassot J, Gouy M, Perriere G, Mouchiroud G (2006) Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Mol Biol Evol 23:1232–1241
Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596
Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domain. Science 241:42–52
Hassel M, Albert K, Hofheinz S (1993) Pattern formation in Hydra vulgaris is controlled by lithium sensitive processes. Dev Biol 156:362–371
Hemmrich G, Anokhin B, Zacharias H, Bosch TC (2007) Molecular phylogenetics in Hydra, a classical model in evolutionary developmental biology. Mol Phylogenetics Evol 44:281–290
Holstein TW, Campbell RD, Tardent P (1990) Identity crisis. Nature 346:21–22
Huang P, Stern MJ (2005) FGF signaling in flies and worms: More and more relevant to vertebrate biology. Cytokine Growth Factor Rev 16:151–158
Imai KS, Hino K, Yagi K, Satoh N, Satou Y (2004) Gene expression profiles of transcription factors and signalling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131(16):4047–4058
Kadam S, McMahon A, Tzou P, Stathopoulos A (2009) FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development 136:739–747
Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298(5601):2157–2167
King N, Carroll SB (2001) A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A 98(26):15032–15037
King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsetn U, Isogai Y, Letunic I, Man M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nicols S, Richter DJ, Salamov A, Sequencing JGI, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tijan R, Grigoriev IV, Rokhsar D (2008) The genome opf the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451(7180):783–788
Kiselyov VV, Li S, Berezin V, Bock E (2009) Insight into the structural mechanism of the bi-modal action of an NCAM mimetic, the C3 peptide. Neurosci Lett 452:224–227
Klämbt C, Glazer L, Shilo BZ (1992) breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev 6:1668–1678
Klingseisen A, Clark I, Gryzik MHA (2009) Differential and overlapping functions of two closely related Drosophila FGF8-like growth factors in mesoderm development. Development 136:2393–2402
Kwiatkowski BA, Kirillova I, Richard RE, Israeli D, Yablonka-Reuveni Z (2008) FGFR4 and its novel splice form in myogenic cells: interplay of glycosylation and tyrosine phosphorylation. J Cell Physiol 215:803–817
Leys SP, Rokhsar DS, Degnan BM (2005) Sponges. Curr Biol 15(4):R114–R115
Li Y, Basilico C, Mansukhani A (1994) Cell transformation by fibroblast growth factors can be suppressed by truncated fibroblast growth factor receptors. Mol Cell Biol 14(11):7660–7669
Liu M, Grigoriev A (2004) Protein domains correlate strongly with exons in multiple eukaryotic genomes—evidence of exon shuffling? Trends Genet 20:399–403
Logsdon JM (2004) Worm genomes hold the smoking guns of intron gain. Proc Natl Acad Sci U S A 101:11195–11196
Martin VJ, Littlefield CL, Archer WE, Bode HR (1997) Embryogenesis in Hydra. Biol Bull 192:345–363
Matus DQ, Thomsen GH, Martindale MQ (2007) FGF signaling in gastrulation and neuronal development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol 217:137–148
Mistry N, Harrington W, Lasda E, Wagner EJ, Garcia-Blanco MA (2003) Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9:209–217
Mohammadi M, Olsen SK, Ibrahimi OA (2005) Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev 16:107–137
Nagendra HG, Harrington AE, Harmer NJ, Pellegrini L, Blundell TL, Burke DF (2001) Sequence analyses and comparative modelling of fly and worm fibroblast growth factor receptors indicate that the determinants for FGF and heparin binding are retained in evolution. FEBS Lett 501:51–58
Neilson KM, Friesel R (1996) Ligand-independent activation of fibroblast growth factor receptors by point mutations in the extracellular, transmembrane and kinase domains. J Biol Chem 271(40):25049–25057
Ogawa K, Kobayashi C, Hayashi T, Orii H, Watanabe K, Agata K (2002) Planarian fibroblast growth factor receptor homologs expressed in stem cells and cephalic ganglions. Dev Growth Differ 44:191–204
Olsen SK, Ibrahimi OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M (2004) Insight into the molecular basis of fibroblast growth factor receptor autoinhibition and ligand binding promiscuity. Proc Natl Acad Sci 101:935–940
Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M (1996) Receptor specificity of the fibroblast growth factor family. J Biol Chem 271:15292–15297
Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Quéinnec E, Da Silva C, Wincker P, Le Guyader H, Leys S, Jackson DJ, Schreiber F, Erpenbeck D, Morgenstern B, Wörheide G, Manuel M (2009) Phylogenomics revives traditional views on deep animal relationships. Curr Biol 19:R339–R341
Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS (2007) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94
Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, Ferrier D, Benes V, de Jong P, Weissenbach J, Bork P, Arendt D (2005) Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science 310:1325–1326
Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U (2008) FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135:1761–1769
Resch A, Xing Y, Alekseyenko A, Modrek B, Lee C (2004) Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res 32:1261–1269
Rokas A, Kruger D, Carroll SB (2005) Animal evolution and the molecular signature of radiation compressed in time. Science 310:1933–1938
Roy SW, Irimia M (2009) Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol Evol 28(8):447–455
Sanchez-Heras E, Howell FV, Williams G, Doherty P (2006) The fibroblast growth factor receptor acidic box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J Biol Chem 281:35208–35216
Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Keisuke U, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, Wiley GB, Macmil SL, Roe BA, Zeller RW, Hastings KEM, Lemaire P, Lindquist E, Endo T, Hotta K, Inaba K (2008) Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into the intron and operon populations. Genome Biology 9(10):R152
Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R (2009) Concatenated analysis sheds light on early metazoan evolution and fuels a modern “Urmetazoon” hypothesis. PLoS Biol 7:e20
Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103:211–225
Schmidt EE, Davies CJ (2007) The origins of polypeptide domains. Bioessays 29:262–270
Schröder R, Beermann A, Wittkopp N, Lutz R (2008) From development to biodiversity—Tribolium castaneum, an insect model organism for short germ band development. Dev Genes Evol 218:119–126
Shimizu A, Tada K, Shukunami C, Shimizu A, Tada K, Shukunami C (2001) A novel alternatively spliced fibroblast growth factor receptor 3 isoform lacking the acid box domain is expressed during chondrogenic differentiation of ATDC5 cells. J Biol Chem 276:11031–11040
Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS (2008) The Trichoplax genome and the nature of placozoans. Nature 454(7207):955–960
Steinberg SF (2008) Structural basis of protein kinase C isoform function. Physiol Rev 88:1341–1378
Sudhop S, Coulier F, Bieller A, Vogt A, Hotz T, Hassel M (2004) Signaling by the FGFR-like tyrosine kinase, Kringelchen, is essential for bud detachment in Hydra vulgaris. Development 131:4001–4011
Sullivan JC, Finnerty JR (2007) A surprising abundance of human disease genes in a simple “basal” animal, the starlet sea anemone (Nematostella vectensis). Genome 50:689–692
Sullivan JC, Reitzel AM, Finnerty JR (2006) A high percentage of introns in human genes were present early in animal evolution: evidence from the basal metazoan Nematostella vectensis. Genome Inform 17:219–229
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ (2005) Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet 21:633–639
Thisse B, Thisse C (2005) Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol 287:390–402
Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955
Wang LJS, Li Y, Paradesi MS, Brown SJ (2007) BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acid Res 35:D476–D479 database issue
Wheelan SJ, Church DM, Ostell JM. (2001) Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 11(11):1952–1957
Acknowledgements
We thank Gabriele Petersen for critical reading of the manuscript and Torsten Rentrop for help with the initial bioinformatic work on the Hydra FGFR gene. We are indebted to Dan Rokhsar and Jarrod Chapman, JGI, for generous access to the unpublished Hydra genome project. We thank the Acorn Worm Genome Sequencing Consortium and the Capitella community for making their data available, which were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community, and the BCM-HGSC for providing the scaffold covering the FGFRs. Anke Beermann and Reinhardt Schröder kindly provided unpublished information concerning Tribolium FGFR. Last but not least, we thank Katja Gessner, our skillful secretary wizardess.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.A. Weisblat
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement 1
Alignment of all used FGFRs and their intron positions superimposed to the protein sequence and domain annotation (PPT 231 kb)
Supplement 2
Assembled and annotated sequences from Trichoplax, Hydra, Nematostella, Lottia Capitella, Tribolium, and Saccoglossus used in the present study (DOC 880 kb)
Supplement 3
Intron size of the three cnidarian FGFRs in comparison to FGFR1 (human) (DOC 61 kb)
Supplement 4
Matrix representation of constitutively spliced introns in FGFR genes across the animal kingdom (DOC 97 kb)
Supplement 5
Intron positions in the highly conserved kinase domains of metazoan receptor tyrosine kinases are not related to intron positions in the choanoflagellate RTK nor in the serine–threonine kinase, PKC (PPT 139 kb)
Supplement 6
Evolutionary relationships of 27 kinases used in this study (PPT 90 kb)
Rights and permissions
About this article
Cite this article
Rebscher, N., Deichmann, C., Sudhop, S. et al. Conserved intron positions in FGFR genes reflect the modular structure of FGFR and reveal stepwise addition of domains to an already complex ancestral FGFR. Dev Genes Evol 219, 455–468 (2009). https://doi.org/10.1007/s00427-009-0309-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-009-0309-5