Abstract
Main Conclusion
This study reveals miRNA indirect regulation of C4 genes in sugarcane through transcription factors, highlighting potential key regulators like SsHAM3a.
Abstract
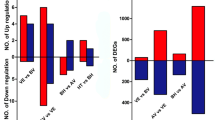
C4 photosynthesis is crucial for the high productivity and biomass of sugarcane, however, the miRNA regulation of C4 genes in sugarcane remains elusive. We have identified 384 miRNAs along the leaf gradients, including 293 known miRNAs and 91 novel miRNAs. Among these, 86 unique miRNAs exhibited differential expression patterns, and we identified 3511 potential expressed targets of these differentially expressed miRNAs (DEmiRNAs). Analyses using Pearson correlation coefficient (PCC) and Gene Ontology (GO) enrichment revealed that targets of miRNAs with positive correlations are integral to chlorophyll-related photosynthetic processes. In contrast, negatively correlated pairs are primarily associated with metabolic functions. It is worth noting that no C4 genes were predicted as targets of DEmiRNAs. Our application of weighted gene co-expression network analysis (WGCNA) led to a gene regulatory network (GRN) suggesting miRNAs might indirectly regulate C4 genes via transcription factors (TFs). The GRAS TF SsHAM3a emerged as a potential regulator of C4 genes, targeted by miR171y and miR171am, and exhibiting a negative correlation with miRNA expression along the leaf gradient. This study sheds light on the complex involvement of miRNAs in regulating C4 genes, offering a foundation for future research into enhancing sugarcane's photosynthetic efficiency.






Similar content being viewed by others
Data availability
The sRNA-Seq reads from the S. spontaneum leaf developmental gradient generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) short read archive (SRA) repository under the accession number PRJNA1030939.
Abbreviations
- DEGs:
-
Differentially expressed genes
- DEmiRNAs:
-
Differentially expressed miRNAs
- FDR:
-
False discovery rate
- GRNs:
-
Gene regulatory networks
- PCC:
-
Pearson correlation coefficient
- TFs:
-
Transcription factors
- TPM:
-
Transcripts per million
References
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107(7):823–826. https://doi.org/10.1016/s0092-8674(01)00616-x
Axtell MJ, Meyers BC (2018) Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 30(2):272–284. https://doi.org/10.1105/tpc.17.00851
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad of Sci USA 102(33):11928–11933. https://doi.org/10.1073/pnas.0505461102
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218(5):683–692. https://doi.org/10.1007/s00425-004-1203-z
Cahoon AB, Takacs EM, Sharpe RM, Stern DB (2008) Nuclear, chloroplast, and mitochondrial transcript abundance along a maize leaf developmental gradient. Plant Mol Biol 66(1–2):33–46. https://doi.org/10.1007/s11103-007-9250-z
Chang Y, Liu W, Shih AC, Shen M, Lu C, Lu MJ, Yang H, Wang T, Chen SCC, Chen SM, Li WH, Ku MSB (2012) Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160(1):165–177. https://doi.org/10.1104/pp.112.203810
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579(26):5923–5931. https://doi.org/10.1016/j.febslet.2005.07.071
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Curaba J, Talbot M, Li Z, Helliwell C (2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13:6. https://doi.org/10.1186/1471-2229-13-6
De Souza AP, Gaspar M, Da Silva EA, Ulian EC, Waclawovsky AJ, Nishiyama MY Jr, Dos Santos RV, Teixeira MM, Souza GM, Buckeridge MS (2008) Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ 31(8):1116–1127. https://doi.org/10.1111/j.1365-3040.2008.01822.x
D’Hont A (2005) Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet Genome Res 109(1–3):27–33. https://doi.org/10.1159/000082378
D’Hont A, Lu YH, León DG, Grivet L, Feldmann P, Lanaud C, Glaszmann JC (1994) A molecular approach to unraveling the genetics of sugarcane, a complex polyploid of the Andropogoneae tribe. Genome 37(2):222–230. https://doi.org/10.1139/g94-031
D’Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC (1996) Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet 250(4):405–413. https://doi.org/10.1007/bf02174028
Ferreira TH, Gentile A, Vilela RD, Costa GGL, Dias LI, Endres L, Menossi M (2012) microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 7(10):e46703. https://doi.org/10.1371/journal.pone.0046703
Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111(6):2367–2372. https://doi.org/10.1073/pnas.1316278111
Ghorai A, Ghosh U (2014) miRNA gene counts in chromosomes vary widely in a species and biogenesis of miRNA largely depends on transcription or post-transcriptional processing of coding genes. Front Genet 5:100. https://doi.org/10.3389/fgene.2014.00100
Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J 40(6):957–967. https://doi.org/10.1111/j.1365-313X.2004.02262.x
Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P (2004) cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16(5):1077–1090. https://doi.org/10.1105/tpc.019729
Hatch MD, Slack CR (1966) Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem J 101(1):103–111. https://doi.org/10.1042/bj1010103
Hibberd JM, Covshoff S (2010) The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol 61(1):181–207. https://doi.org/10.1146/annurev-arplant-042809-112238
Hu W, Hua X, Zhang Q, Wang J, Shen Q, Zhang X, Wang K, Yu Q, Lin Y-R, Ming R, Zhang J (2018) New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics. BMC Plant Biol 18(1):270. https://doi.org/10.1186/s12870-018-1495-y
Hwang D-G, Park JH, Lim JY, Kim D, Choi Y, Kim S, Reeves G, Yeom S-I, Lee J-S, Park M, Kim S, Choi I-Y, Choi D, Shin C (2013) The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding microRNA functional roles in hot pepper. PLoS ONE 8(5):e64238. https://doi.org/10.1371/journal.pone.0064238
Jatan R, Chauhan PS, Lata C (2020) High-throughput sequencing and expression analysis suggest the involvement of Pseudomonas putida RA-responsive micrornas in growth and development of Arabidopsis. Int J of Mol Sci 21(15):5468. https://doi.org/10.3390/ijms21155468
Jiang Q, Hua X, Shi H, Liu J, Yuan Y, Li Z, Li S, Zhou M, Yin C, Dou M, Qi N, Wang Y, Zhang M, Ming R, Tang H, Zhang J (2023) Transcriptome dynamics provides insights into divergences of the photosynthesis pathway between Saccharum officinarum and Saccharum spontaneum. Plant J 113(6):1278–1294. https://doi.org/10.1111/tpj.16110
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37(8):907–915. https://doi.org/10.1038/s41587-019-0201-4
Lai X, Stigliani A, Vachon G, Carles C, Smaczniak C, Zubieta C, Kaufmann K, Parcy F (2019) Building transcription factor binding site models to understand gene regulation in plants. Mol Plant 12(6):743–763. https://doi.org/10.1016/j.molp.2018.10.010
Li A, Mao L (2007) Evolution of plant microRNA gene families. Cell Res 17(3):212–218. https://doi.org/10.1038/sj.cr.7310113
Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T, Brutnell TP (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42(12):1060–1067. https://doi.org/10.1038/ng.703
Li M, Liang Z, He S, Zeng Y, Jing Y, Fang W, Wu K, Wang G, Ning X, Wang L, Li S, Tan H, Tan F (2017) Genome-wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.). BMC Genom 18(1):754. https://doi.org/10.1186/s12864-017-4053-3
Li Z, Hua X, Zhong W, Yuan Y, Wang Y, Wang Z, Ming R, Zhang J (2020) Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol 61(3):616–630. https://doi.org/10.1093/pcp/pcz227
Li G, Chen C, Chen P, Meyers BC, Xia RJSB (2023) sRNAminer: a multifunctional toolkit for next-generation sequencing small RNA data mining in plants. Sci Bull S2095–9273(23):00935–01930. https://doi.org/10.1016/j.scib.2023.12.049
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14(7):1605–1619. https://doi.org/10.1105/tpc.003210
Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29(3):315–330. https://doi.org/10.1111/j.1365-3040.2005.01493.x
Loudya N, Mishra P, Takahagi K, Uehara-Yamaguchi Y, Inoue K, Bogre L, Mochida K, López-Juez E (2021) Cellular and transcriptomic analyses reveal two-staged chloroplast biogenesis underpinning photosynthesis build-up in the wheat leaf. Genome Biol 22(1):151. https://doi.org/10.1186/s13059-021-02366-3
Ma Z, Hu X, Cai W, Huang W, Zhou X, Luo Q, Yang H, Wang J, Huang J (2014) Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet 10(8):e1004519. https://doi.org/10.1371/journal.pgen.1004519
Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, Turgeon R, van Wijk KJ (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22(11):3509–3542. https://doi.org/10.1105/tpc.110.079764
Marchiori PER, Ribeiro RV, da Silva L, Machado RS, Machado EC, Scarpari MS (2010) Plant growth, canopy photosynthesis and light availability in three sugarcane varieties. Sugar Tech 12(2):160–166. https://doi.org/10.1007/s12355-010-0031-7
Marchiori PER, Machado EC, Ribeiro RV (2014) Photosynthetic limitations imposed by self-shading in field-grown sugarcane varieties. Field Crops Res 155:30–37. https://doi.org/10.1016/j.fcr.2013.09.025
Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PMJFl (2006) The evolution and diversification of dicers in plants. FEBS Lett 580(10):2442–2450. https://doi.org/10.1016/j.febslet.2006.03.072
Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98(22):12826–12831. https://doi.org/10.1073/pnas.221252798
Mochizuki N, Tanaka R, Grimm B, Masuda T, Moulin M, Smith AG, Tanaka A, Terry MJ (2010) The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 15(9):488–498. https://doi.org/10.1016/j.tplants.2010.05.012
Moore PH (1995) Temporal and spatial regulation of sucrose accumulation in the sugarcane stem. Aust J Plant Physiol 22(4):661–680. https://doi.org/10.1071/PP9950661
Pan J, Huang D, Guo Z, Kuang Z, Zhang H, Xie X, Ma Z, Gao S, Lerdau MT, Chu C, Li L (2018) Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J Integr Plant Biol 60(4):323–340. https://doi.org/10.1111/jipb.12634
Pei LL, Zhang LL, Liu X, Jiang J (2023) Role of microRNA miR171 in plant development. PeerJ 11:e15632. https://doi.org/10.7717/peerj.15632
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. https://doi.org/10.1038/nbt.3122
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT. StringTie and Ballgown Nat Protoc 11(9):1650–1667. https://doi.org/10.1038/nprot.2016.095
Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, Weber AP (2011) Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23(12):4208. https://doi.org/10.1105/tpc.111.090324
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110(4):513–520. https://doi.org/10.1016/s0092-8674(02)00863-2
Schulze S, Schäfer BN, Parizotto EA, Voinnet O, Theres K (2010) LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J 64(4):668–678. https://doi.org/10.1111/j.1365-313X.2010.04359.x
Sharpe RM, Mahajan A, Takacs EM, Stern DB, Cahoon AB (2011) Developmental and cell type characterization of bundle sheath and mesophyll chloroplast transcript abundance in maize. Curr Genet 57(2):89–102. https://doi.org/10.1007/s00294-010-0329-8
Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11(1):431. https://doi.org/10.1186/1471-2164-11-431
Tian F, Yang D-C, Meng Y-Q, Jin J, Gao G (2020) PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res 48(D1):D1104–D1113. https://doi.org/10.1093/nar/gkz1020
Wai CM, VanBuren R, Zhang J, Huang L, Miao W, Edger PP, Yim WC, Priest HD, Meyers BC, Mockler T, Smith JAC, Cushman JC, Ming R (2017) Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. Plant J 92(1):19–30. https://doi.org/10.1111/tpj.13630
Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ (2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3(5):794–806. https://doi.org/10.1093/mp/ssq042
Wang L, Peterson RB, Brutnell TP (2011) Regulatory mechanisms underlying C4 photosynthesis. New Phytol 190(1):9–20. https://doi.org/10.1111/j.1469-8137.2011.03649.x
Wang L, Czedik-Eysenberg A, Mertz RA, Si Y, Tohge T, Nunes-Nesi A, Arrivault S, Dedow LK, Bryant DW, Zhou W, Xu J, Weissmann S, Studer A, Li P, Zhang C, LaRue T, Shao Y, Ding Z, Sun Q, Patel RV, Turgeon R, Zhu X, Provart NJ, Mockler TC, Fernie AR, Stitt M, Liu P, Brutnell TP (2014) Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat Biotechnol 32(11):1158–1165. https://doi.org/10.1038/nbt.3019
Wu HJ, Wang ZM, Wang M, Wang XJ (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161(4):1875–1884. https://doi.org/10.1104/pp.113.215962
Xu J, Li Y, Wang Y, Liu X, Zhu X-G (2017) Altered expression profiles of microRNA families during de-etiolation of maize and rice leaves. BMC Res Notes 10(1):108. https://doi.org/10.1186/s13104-016-2367-x
Zhang JP, Yu Y, Feng YZ, Zhou YF, Zhang F, Yang YW, Lei MQ, Zhang YC, Chen YQ (2017) MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol 175(3):1175–1185. https://doi.org/10.1104/pp.17.01169
Zhang J, Zhang X, Tang H, Zhang Q, Hua X, Ma X, Zhu F, Jones T, Zhu X et al (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50(11):1565–1573. https://doi.org/10.1038/s41588-018-0237-2
Zhang Q, Qi Y, Pan H, Tang H, Wang G, Hua X, Wang Y, Lin L, Li Z, Li Y, Yu F, Yu Z, Huang Y, Wang T, Ma P, Dou M, Sun Z, Wang Y, Wang H, Zhang X, Yao W, Wang Y, Liu X, Wang M, Wang J, Deng Z, Xu J, Yang Q, Liu Z, Chen B, Zhang M, Ming R, Zhang J (2022) Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat Genet 54(6):885–896. https://doi.org/10.1038/s41588-022-01084-1
Acknowledgements
We thank the support of the National Key Research and Development program (2021YFF1000101 and 2021YFF1000104), the National Natural Science Foundation of China (32272196), the Sugarcane Research Foundation of Guangxi University (Grant No.2022GZB007), and the fellowship of China Postdoctoral Science Foundation (2022MD723761). We also thank Chengjie Chen and Guanliang Li at the South China Agricultural University for their useful advice and excellent technical assistance.
Author information
Authors and Affiliations
Contributions
JZ conceived and designed research. XH, MD, DZ, HS, YL, QW, and SL conducted experiments. XH, MD, ZL, YZ, RG, YH, YQ, BW and QiyunW analyzed data. XH, ZL and MD wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2024_4390_MOESM6_ESM.xlsx
Dataset S6 List of transcription factors included in modules co-expressed with C4 genes and targeted by miRNAs used for the construction of gene regulatory networks
425_2024_4390_MOESM7_ESM.xlsx
Dataset S7 Details of transcription factors with correlations with miRNAs in the shared miRNA-TF module in GRN1 and GRN2
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hua, X., Li, Z., Dou, M. et al. Transcriptome and small RNA analysis unveils novel insights into the C4 gene regulation in sugarcane. Planta 259, 120 (2024). https://doi.org/10.1007/s00425-024-04390-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04390-6