Abstract
Main Conclusion
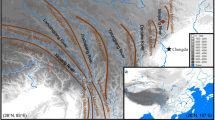
The divergence of subsect. Gerardianae was likely triggered by the uplift of the Qinghai–Tibetan Plateau and adjacent mountains. Pinus bungeana might have probably experienced expansion since Last Interglacial period.
Abstract
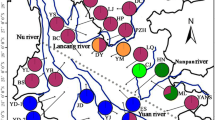
Historical geological and climatic oscillations have profoundly affected patterns of nucleotide variability, evolutionary history, and species divergence in numerous plants of the Northern Hemisphere. However, how long-lived conifers responded to geological and climatic fluctuations in East Asia remain poorly understood. Here, based on paternally inherited chloroplast genomes and maternally inherited mitochondrial DNA markers, we investigated the population demographic history and molecular evolution of subsect. Gerardianae (only including three species, Pinus bungeana, P. gerardiana, and P. squamata) of Pinus. A low level of nucleotide diversity was found in P. bungeana (π was 0.00016 in chloroplast DNA sequences, and 0.00304 in mitochondrial DNAs). The haplotype-based phylogenetic topology and unimodal distributions of demographic analysis suggested that P. bungeana probably originated in the southern Qinling Mountains and experienced rapid population expansion since Last Interglacial period. Phylogenetic analysis revealed that P. gerardiana and P. squamata had closer genetic relationship. The species divergence of subsect. Gerardianae occurred about 27.18 million years ago (Mya) during the middle to late Oligocene, which was significantly associated with the uplift of the Qinghai–Tibetan Plateau and adjacent mountains from the Eocene to the mid-Pliocene. The molecular evolutionary analysis showed that two chloroplast genes (psaI and ycf1) were under positive selection, the genetic lineages of P. bungeana exhibited higher transition and nonsynonymous mutations, which were involved with the strongly environmental adaptation. These findings shed light on the population evolutionary history of white pine species and provide striking insights for comprehension of their species divergence and molecular evolution.







Similar content being viewed by others
Data availability
The authors do not have permission to share data.
Abbreviations
- cpDNA:
-
Chloroplast DNA
- cp genome:
-
Chloroplast genome
- mtDNA:
-
Mitochondrial DNA
- Mya:
-
Million years ago
- QTP:
-
Qinghai–Tibetan Plateau
- RMSD:
-
Root-mean-square deviation
- SNP:
-
Single-nucleotide polymorphism
References
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bolger AM, Logse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120
Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB (2004) Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc Natl Acad Sci USA 101:15255–15260
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159
Crampton-Platt A, Timmermans MJ, Gimmel ML, Kutty SN, Cockerill TD, Khen CV, Vogler AP (2015) Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a bornean rainforest sample. Mol Biol Evol 32:2302–2316
Denisov G, Walenz B, Halpern AL, Miller J, Axelrod N, Levy S, Sutton G (2008) Consensus generation and variant detection by celera assembler. Bioinformatics 24:1035–1040
Ding L, Liao JC, Liu NF (2020) The uplift of the Qinghai-Tibet Plateau and glacial oscillations triggered the diversification of Tetraogallus (Galliformes, Phasianidae). Ecol Evol 10:1722–1736
Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res 47:10543–10552
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from plant tissue. Focus 12:13–15
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with beauti and the beast 1.7. Mol Biol Evol 29:1969–1973
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Eveno E, Collada C, Guevara MA, Léger V, Soto A, Díaz L, Léger P, González-Martínez SC, Cervera MT, Plomin C, Garnier-Géré PH (2008) Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol Biol Evol 25:417–437
Excoffier L, Laval G, Schneider S (2007) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fay JC, Wu CI (2000) Hitchhiking under positive darwinian selection. Genetics 155:1405–1413
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fu LG, Li N, Mill RR (1999) Pinaceae. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China. Science Press, Missouri Botanical Garden Press, Beijing, St. Louis, pp 25–32
Gernandt DS, Liston A, Piñero D (2003) Phylogenetics of Pinus subsections Cembroides and Nelsoniae inferred from cpDNA sequences. Syst Bot 28:657–673
Gernandt DS, López GG, García SO, Liston A (2005) Phylogeny and classification of Pinus. Taxon 54:29–42
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89:415–426
Guo J-F, Wang BS, Liu Z-L, Mao JF, Wang XR, Zhao W (2021) Low genetic diversity and population connectivity fuel vulnerability to climate change for the Tertiary relict pine Pinus bungeana. J Syst Evol 61:143–156
Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P (2005) Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res 15:790–799
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-abaiting and iterative mapping approach. Nucleic Acids Res 41:e129
Hall D, Olsson J, Zhao W, Kroon J, Wennström U, Wang XR (2021) Divergent patterns between phenotypic and genetic variation in Scots pine. Plant Commun 2:100139
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Hao ZZ, Liu YY, Nazaire M, Wei XX, Wang XQ (2015) Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): implications for Northern Hemisphere biogeography. Mol Phylogenet Evol 87:65–79
Hu CY, Zheng XY, Teng YW (2011) Characterization and phylogenetic utility of non-coding chloroplast regions trnL-trnF and accD-psaI in Pyrus. Acta Horticulturae Sinica 38:2261–2272
Huang JH, Huang JH, Liu CR, Zhang JL, Lu XH, Ma KP (2016) Diversity hotspots and conservation gaps for the Chinese endemic seed flora. Biol Conserv 198:104–112
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
IUCN (2012) Guidelines for application of IUCN red list criteria at regional and national levels: Version 4.0. IUCN
Jiang XZ, Yu HY, Xiang MC, Liu XY, Liu XZ (2011) Echinochlamydosporium variabile, a new genus and species of Zygomycota from soil nematodes. Fungal Divers 46:43–51
Jin W-T, Gernandt DS, Wehenkel C, Xia XM, Wei XX, Wang XQ (2021) Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc Natl Acad Sci USA 118:e2022302118
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software versions 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Stur-rock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Krutovsky KV, Neale DB (2005) Nucleotide diversity and linkage disequilibrium in cold-hardiness- and wood quality-related candiate genes in Douglas fir. Genetics 171:2029–2041
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Li Y, Stocks M, Hemmilä S, Källman T, Zhu HT, Zhou Y, Chen J, Liu JQ, Lascoux M (2009) Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Mol Biol Evol 27:1001–1014
Li Y, Gao QB, Gengji ZM, Jia L-K, Wang Z-H, Chen S-L (2018) Rapid intraspecific diversification of the alpine species Saxifraga sinomontana (Saxifragaceae) in the Qinghai-Tibetan Plateau and Himalayas. Front Genet 9:381
Librado P, Rozas J (2009) Dnasp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu YY, Jin WT, Wei XX, Wang XQ (2019) Cryptic speciation in the Chinese white pine (Pinus armandii): implications for the high species diversity of conifers in the Hengduan Mountains, a global biodiversity hotspot. Mol Phylogenet Evol 138:114–125
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Ma XF, Szmidt AE, Wang XR (2006) Genetic structure and evolutionary history of a diploid hybrid pine Pinus densata inferred from the nucleotide variation at seven gene loci. Mol Biol Evol 23:807–816
Meng ZL, Zaykin DV, Xu CF, Wagner M, Ehm MG (2003) Selection of genetic markers for association analyses, using linkage disequilibrium and haplotypes. Am J Hum Genet 73:115–130
Mitchell C (1993) MultAlin–multiple sequence alignment. Bioinformatics 9:614
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304
Ogawa T, Ishii C, Kagawa D, Muramoto K, Kamiya H (1999) Accelerated evolution in the protein-coding region of galectin cDNAs, congerin I and congerin II, from skin mucus of conger eel (Conger myriaster). Biosci Biotechnol Biochem 63:1203–1208
Palmer JD, Herbon LA (1988) Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol 28:87–97
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Potter KM, Hipkins VD, Mahalovich MF, Means RE (2015) Nuclear genetic variation across the range of ponderosa pine (Pinus ponderosa): phylogeographic, taxonomic and conservation implications. Tree Genet Genomes 11:38
Price RA, Liston A, Strauss SH (1998) Phylogeny and systematics of Pinus. In: Richardson D (ed) Ecology and biogeography of Pinus. Cambridge University Press, Cambridge, pp 49–68
Pyhäjärvi T, Salmela MJ, Savolainen O (2008) Colonization routes of Pinus sylvestris inferred from distribution of mitochondrial DNA variation. Tree Genet Genom 4:247–254
Qian H, Ricklefs RE (2000) Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407:180–182
Rodríguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H (2007) Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol 56:389–399
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Saitou N, Ueda S (1994) Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol Biol Evol 11:504–512
Sloan DB, Taylor DR (2012) Evolutionary rate variation in organelle genomes: the role of mutational processes. In: Bullerwell CE (ed) Organelle genetics. Springer, Berlin Heidelberg, pp 123–146
Sobel JM, Chen GF, Watt LR, Schemske DW (2010) The biology of speciation. Evolution 64:295–315
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Straub SCK, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A (2012) Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot 99:349–364
Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics 123:597–601
Twyford AD, Ness RW (2017) Strategies for complete plastid genome sequencing. Mol Ecol Resour 17:858–868
Umar Z, Dong WL, Zhang TT, Wang R-N, Shahzad K, Ma X-F, Li Z-H (2020) Comparative plastid genomics of Pinus species: insights into sequence variations and phylogenetic relationships. J Syst Evol 58:118–132
Walker JF, Walker-Hale N, Vargas OM, Larson DA, Stull GW (2019) Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7:e7747
Wang B, Mao JF, Gao J, Zhao W, Wang XR (2011) Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata. Mol Ecol 20:3796–3811
Wang S, Shi C, Gao LZ (2013) Plastid genome sequence of a wild woody oil species, Prinsepia utilis, provides insights into evolutionary and mutational patterns of Rosaceae chloroplast genomes. PLoS One 8:e73946
Wang W, Chen X, Wolin SL, Xiong Y (2018a) Structural basis for tRNA mimicry by a bacterial Y RNA. Structure 26:1635–1644
Wang YC, Liao L, Li ZZ (2018b) Genetic differentiation of Actinidia chinensis and analysis of gene flow barriers in the Qinling Mountains, the species’ northern distribution boundary. Genet Resour Crop Evol 65:881–895
Wang N, Dong W-L, Zhang X-J, Zhou T, Huang X-J, Li B-G, Liu J-N, Ma X-F, Li Z-H (2021) Evolutionary characteristics and phylogeny of cotton chloroplast tRNAs. Planta 254:116
Weber F, Dietrich A, Weil J-H, Maréchal-Drouard L (1990) A potato mitochondrial isoleucine tRNA is coded for by a mitochondrial gene possessing a methionine anticodon. Nucl Acids Res 18:5027–5030
Wu SD, Wang Y, Wang ZF, Shrestha N, Liu JQ (2022) Species divergence with gene flow and hybrid speciation on the Qinghai–Tibet Plateau. New Phytol 234:392–404
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917
Yang YX, Zhi LQ, Jia Y, Zhong Q-Y, Liu Z-L, Yue M, Li Z-H (2020) Nucleotide diversity and demographic history of Pinus bungeana, an endangered conifer species endemic in China. J Syst Evol 58:282–294
Yin A, Harrison TM (2000) Geologic evolution of the Himalayan–Tibetan orogen. Annu Rev Earth Planet Sci 28:211–280
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693
Zhang ZY, Yang JB, Li DZ (2003) Phylogenetic relationship of an extremely endangered species, Pinus squamata (Pinaceae) inferred from four sequences of the chloroplast genome and ITS of the nuclear ribosomal DNA. Acta Bot Sin 45:530–535
Zhang TT, Liu H, Gao QY, Yang T, Liu JN, Ma XF, Li Z-H (2020a) Gene transfer and nucleotide sequence evolution by Gossypium cytoplasmic genomes indicates novel evolutionary characteristics. Plant Cell Rep 39:765–777
Zhang Y, Wang J, Xiao Y (2020b) 3dRNA: Building RNA 3D structure with improved template library. Comput Struct Biotechnol J 18:2416–2423
Zhao W, Sun YQ, Pan J, Sullivan AR, Arnold ML, Mao JF, Wang XR (2020) Effects of landscapes and range expansion on population structure and local adaptation. New Phytol 228:330–343
Zhong QY, Fu XG, Zhang TT, Zhou T, Yue M, Liu JN, Li ZH (2021) Phylogeny and evolution of chloroplast tRNAs in Adoxaceae. Ecol Evol 11:1294–1309
Zhou YF, Zhang LR, Liu JQ, Wu GL, Savolainen O (2014) Climatic adaptation and ecological divergence between two closely related pine species in Southeast China. Mol Ecol 23:3504–3522
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31970359) and the Key Program of Research and Development of Shaanxi Province (2022ZDLSF06-02).
Author information
Authors and Affiliations
Contributions
ZHL and YXK designed the research and contributed the materials. JXQ and ASY performed the experiments. TTZ and CLY carried out data analysis. MLL contributed analysis tools. TTZ wrote the first draft of the manuscript. ZHL revised the paper. All authors have approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2023_4316_MOESM4_ESM.pdf
Fig. S4 LnP(D) and ΔK test based on the cpDNA sequences for the 18 populations of P. bungeana. a LnP(D) for K from 1 to 15. b ΔK for K from 1 to 15 (PDF 123 kb)
425_2023_4316_MOESM5_ESM.pdf
Fig. S5 Results of structure based on the cpDNA sequences for the 18 populations of P. bungeana (when K was 2, 3, and 4) (PDF 353 kb)
425_2023_4316_MOESM6_ESM.pdf
Fig. S6 LnP(D) and ΔK test based on the mtDNA sequences for the 18 populations of P. bungeana. a LnP(D) for K from 1 to 15. b ΔK for K from 1 to 15 (PDF 125 kb)
425_2023_4316_MOESM7_ESM.pdf
Fig. S7 Results of structure based on the mtDNA sequences for the 18 populations of P. bungeana (when K was 2, 3, and 4) (PDF 365 kb)
425_2023_4316_MOESM8_ESM.pdf
Fig. S8 Secondary structure of typical clover and unconventional structures for chloroplast genomic tRNAs. a The typical clover structure for tRNAArg. b tRNASer with unconventional structure. c tRNALeu with unconventional structure. d tRNATyr with unconventional structure (PDF 408 kb)
425_2023_4316_MOESM9_ESM.pdf
Fig. S9 Tertiary structures of typical and unconventional chloroplast genomic tRNAs. a The typical tertiary structure for tRNAArg. b tRNASer with unconventional tertiary structure. c tRNALeu with unconventional tertiary structure. d tRNATyr with unconventional tertiary structure (PDF 20991 kb)
425_2023_4316_MOESM10_ESM.pdf
Fig. S10 The root-mean-square deviation (RMSD) of the same tRNAs molecules between P. bungeana (red) and P. squamata (blue). a The RMSD of tRNAArg (0.172) between P. bungeana (red) and P. squamata (blue). b The RMSD of tRNATrp (1.748) between P. bungeana (red) and P. squamata (blue). c The RMSD of tRNAMet (10.542) between P. bungeana (red) and P. squamata (blue). d The RMSD of tRNAGlu (0.578) between P. bungeana (red) and P. squamata (blue) (PDF 6516 kb)
425_2023_4316_MOESM11_ESM.pdf
Fig. S11 The root-mean-square deviation (RMSD) of the same tRNAs molecules between P. bungeana (red) and P. gerardiana (blue). a The RMSD of tRNAArg (0.185) between P. bungeana (red) and P. gerardiana (blue). b The RMSD of tRNATrp (0.018) between P. bungeana (red) and P. gerardiana (blue). c The RMSD of tRNAMet (8.172) between P. bungeana (red) and P. gerardiana (blue). d The RMSD of tRNAGlu (0.297) between P. bungeana (red) and P. gerardiana (blue) (PDF 7341 kb)
425_2023_4316_MOESM12_ESM.pdf
Fig. S12 The root-mean-square deviation (RMSD) of the same tRNA molecules between P. squamata (red) and P. gerardiana (blue). a The RMSD of tRNAArg (0.036) between P. squamata (red) and P. gerardiana (blue). b The RMSD of tRNATrp (1.734) between P. squamata (red) and P. gerardiana (blue). c The RMSD of tRNAMet (8.173) between P. squamata (red) and P. gerardiana (blue). d The RMSD of tRNAGlu (0.571) between P. squamata (red) and P. gerardiana (blue) (PDF 6315 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, TT., Yan, CL., Qiao, JX. et al. Demographic dynamics and molecular evolution of the rare and endangered subsect. Gerardianae of Pinus: insights from chloroplast genomes and mitochondrial DNA markers. Planta 259, 45 (2024). https://doi.org/10.1007/s00425-023-04316-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04316-8