Abstract
Main conclusion
Multiplexed Cas9-based genome editing of cotton resulted in reduction of viral load with asymptomatic cotton plants. In depth imaging of proteomic dynamics of resulting CLCuV betasatellite and DNA-A protein was also performed.
Abstract
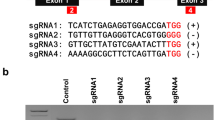
The notorious cotton leaf curl virus (CLCuV), which is transmitted by the sap-sucking insect whitefly, continuously damages cotton crops. Although the application of various toxins and RNAi has shown some promise, sustained control has not been achieved. Consequently, CRISPR_Cas9 was applied by designing multiplex targets against DNA-A (AC2 and AC3) and betasatellite (βC1) of CLCuV using CRISPR direct and ligating into the destination vector of the plant using gateway ligation method. The successful ligation of targets into the destination vector was confirmed by the amplification of 1049 bp using a primer created from the promoter and target, while restriction digestion using the AflII and Asc1 enzymes determined how compact the plasmid developed and the nucleotide specificity of the plasmid was achieved through Sanger sequencing. PCR confirmed the successful introduction of plasmid into CKC-1 cotton variety. Through Sanger sequencing and correlation with the mRNA expression of DNA-A and betasatellite in genome-edited cotton plants subjected to agroinfiltration of CLCuV infectious clone, the effectiveness of knockout was established. The genome-edited cotton plants demonstrated edited efficacy of 72% for AC2 and AC3 and 90% for the (βC1) through amplicon sequencing, Molecular dynamics (MD) simulations were used to further validate the results. Higher RMSD values for the edited βC1 and AC3 proteins indicated functional loss caused by denaturation. Thus, CRISPR_Cas9 constructs can be rationally designed using high-throughput MD simulation technique. The confidence in using this technology to control plant virus and its vector was determined by the knockout efficiency and the virus inoculation assay.
Graphical abstract









Similar content being viewed by others
Data availability
The sequences of CLCuV were retrieved from the nucleotide domain of NCBI website (https://www.ncbi.nlm.nih.gov/nucleotide/) with accession numbers of KY992859.1, MK370435.1, HE601939.1. The study does not involve the isolation of any new gene. Instead, it involves CRISPR_Cas-mediated knockdown of viral genes into the local cotton variety.
References
Ali A, Ahmed Z, Guo Z (2021) 4 cotton growing in Pakistan. Pest Management in Cotton: A Global Perspective 53
Asif M, Muhammad R, Akbar W, Toufique M (2017) Response of various cotton genotypes against sucking and bollworm complexes. Pakistan J Agric Agric Eng Vet Sci 33(1):37–45
Azam S, Rao AQ, Shad M, Ahmed M, Gul A, Latif A, Ali MA, Husnain T, Shahid AA (2021) Development of broad-spectrum and sustainable resistance in cotton against major insects through the combination of Bt and plant lectin genes. Plant Cell Rep 40(4):707–721
Bakhsh A, Rao AQ, Shahid AA, Husnain T, Riazuddin S (2009) Insect resistance and risk assessment studies in advance lines of Bt cotton harboring Cry1Ac and Cry2A genes. Am Eurasian J Agric Environ Sci 6(1):1–11
Bakhsh A, Anayol E, Özcan SF, Hussain T, Aasim M, Khawar KM, Özcan S (2015) An insight into cotton genetic engineering (Gossypium hirsutum L.): current endeavors and prospects. Acta Physiol Plant 37(8):1–17
Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I (2013) High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 8(3):539–554
Cerutti H, Ma X, Msanne J, Repas T (2011) RNA-mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryot Cell 10(9):1164–1172
Cramer P (2021) AlphaFold2 and the future of structural biology. Nat Struct Mol Biol 28(9):704–705
DeLano WL (2002) Pymol: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40(1):82–92
Gajula MP, Kumar A, Ijaq J (2016) Protocol for molecular dynamics simulations of proteins. Bio-Protoc 6(23):e2051–e2051
Gao W, Long L, Tian X, Xu F, Liu J, Singh PK, Botella JR, Song C (2017) Genome editing in cotton with the CRISPR/Cas9 system. Front Plant Sci 8:1364
Gul A, Hussain G, Iqbal A, Rao AQ, Yasmeen A, Shahid N, Ahad A, Latif A, Azam S, Samiullah TR (2020) Constitutive expression of Asparaginase in Gossypium hirsutum triggers insecticidal activity against Bemisia tabaci. Sci Rep 10(1):1–11
Hasley JAR, Navet N, Tian M (2021) CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PLoS One 16(6):e0253245
Hong JK, Suh EJ, Park SR, Park J, Lee Y-H (2021) Multiplex CRISPR/Cas9 mutagenesis of BrVRN1 delays flowering time in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Agriculture 11(12):1286
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832
Hu D, Yu Y, Wang C, Long Y, Liu Y, Feng L, Lu D, Liu B, Jia J, Xia R (2021) Multiplex CRISPR-Cas9 editing of DNA methyltransferases in rice uncovers a class of non-CG methylation specific for GC-rich regions. Plant Cell 33(9):2950–2964
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589
Kataria SK, Pal RK, Kumar V, Singh P (2019) Population dynamics of whitefly, Bemisia tabaci (Gennadius), as influenced by weather conditions infesting Bt cotton hybrid. J Agrometeorol 21(4):504–509
Karplus M, Kuriyan J (2005) Molecular dynamics and protein function. Proc Nat Acad Sci 102(19):6679–6685
Khan ZA, Abdin MZ, Khan JA (2015) Functional characterization of a strong bi-directional constitutive plant promoter isolated from cotton leaf curl Burewala virus. PLoS One 10(3):e0121656
Khan AA, El-Sayed A, Akbar A, Mangravita-Novo A, Bibi S, Afzal Z, Norman DJ, Ali GS (2017) A highly efficient ligation-independent cloning system for CRISPR/Cas9 based genome editing in plants. Plant Methods 13(1):1–13
Kheirodin A, Simmons AM, Legaspi JC, Grabarczyk EE, Toews MD, Roberts PM, Chong J-H, Snyder WE, Schmidt JM (2020) Can generalist predators control Bemisia tabaci? InSects 11(11):823
Kumar V, AlMomin S, Rahman MH, Shajan A (2020) Use of CRISPR in climate smart/resilient agriculture. In: CRISPR/Cas genome editing. Springer, pp 131–164
Kutnjak D, Tamisier L, Adams I, Boonham N, Candresse T, Chiumenti M, De Jonghe K, Kreuze JF, Lefebvre M, Silva G (2021) A primer on the analysis of high-throughput sequencing data for detection of plant viruses. Microorganisms 9(4):841
Lee JH, Mazarei M, Pfotenhauer AC, Dorrough AB, Poindexter MR, Hewezi T, Lenaghan SC, Graham DE, Stewart CN Jr (2020) Epigenetic footprints of CRISPR/Cas9-mediated genome editing in plants. Front Plant Sci 10:1720
Lei Y, Lu L, Liu H-Y, Li S, Xing F, Chen L-L (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7(9):1494–1496
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Mahmood TB, Hossan MI, Mahmud S, Shimu MSS, Alam MJ, Bhuyan MMR, Emran TB (2022) Missense mutations in spike protein of SARS‐CoV‐2 delta variant contribute to the alteration in viral structure and interaction with hACE2 receptor. Immun Inflamm Dis 10(9):e683
Majid M, Andleeb S (2019) Designing a multi-epitopic vaccine against the enterotoxigenic Bacteroides fragilis based on immunoinformatics approach. Sci Rep 9(1):1–15
Matsuo K, Atsumi G (2019) CRISPR/Cas9-mediated knockout of the RDR6 gene in Nicotiana benthamiana for efficient transient expression of recombinant proteins. Planta 250(2):463–473
Misaka BC, Wosula EN, Marchelo-d’Ragga PW, Hvoslef-Eide T, Legg JP (2020) Genetic diversity of Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae) colonizing sweet potato and cassava in South Sudan. InSects 11(1):58
Mubarik MS, Wang X, Khan SH, Ahmad A, Khan Z, Amjid MW, Razzaq MK, Ali Z, Azhar MT (2021) Engineering broad-spectrum resistance to cotton leaf curl disease by CRISPR-Cas9 based multiplex editing in plants. GM Crops Food 12(2):647–658
Mubin M, Hussain M, Briddon RW, Mansoor S (2011) Selection of target sequences as well as sequence identity determine the outcome of RNAi approach for resistance against cotton leaf curl geminivirus complex. Virol J 8(1):1–8
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7(1):1–6
Paredes-Montero JR, Hameed U, Zia-Ur-Rehman M, Rasool G, Haider MS, Herrmann H-W, Brown JK (2019) Demographic expansion of the predominant Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae) mitotypes associated with the Cotton leaf curl virus epidemic in Pakistan. Ann Entomol Soc Am 112(3):265–280
Paredes-Montero JR, Haq Q, Mohamed AA, Brown JK (2021) Phylogeographic and SNPs analyses of Bemisia tabaci B mitotype populations reveal only two of eight haplotypes are invasive. Biology 10(10):1048
Reece-Hoyes JS, Walhout AJ (2018) Gateway recombinational cloning. Cold Spring Harb Protoc 2018(1):pdb. top094912
Salisu IB, Shahid AA, Yaqoob A, Gul A, Rao AQ (2021) Evaluation the effect of subchronic feeding of transgenic cotton line (CKC1) on the faecal microbiota of albino rabbits. J Anim Physiol Anim Nutr 105(2):354–363
Schachtsiek J, Stehle F (2019) Nicotine-free, nontransgenic tobacco (Nicotiana tabacum L.) edited by CRISPR-Cas9. Plant Biotechnol J 17(12):2228
Shad M, Yasmeen A, Azam S, Bakhsh A, Latif A, Shahid N, Sadaqat S, Rao AQ, Shahid AA (2021) Enhancing the resilience of transgenic cotton for insect resistance. Mol Biol Reports 49(6):5315–5323
Shah H, Khan K, Khan N, Badshah Y, Ashraf NM, Shabbir M (2022) Impact of deleterious missense PRKCI variants on structural and functional dynamics of protein. Sci Rep 12(1):1–17
Sher Z, Majid MU, Hassan S, Batool F, Aftab B, Rashid B (2021) Identification, isolation and characterization of GaCyPI gene in Gossypium arboreum under cotton leaf curl virus disease stress. Phyton 90(6):1613
Shukla N, Verma S, Babu GS, Saxena S (2017) Strategy for generic resistance against begomoviruses through RNAi. In: Begomoviruses: occurrence and management in Asia and Africa. Springer, pp 137–155
Tennakoon C, Purbojati RW, Sung W-K (2012) BatMis: a fast algorithm for k-mismatch mapping. Bioinformatics 28(16):2122–2128
Uniyal AP, Yadav SK, Kumar V (2019) The CRISPR–Cas9, genome editing approach: a promising tool for drafting defense strategy against begomoviruses including cotton leaf curl viruses. J Plant Biochem Biotechnol 28(2):121–132
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci 112(11):3570–3575
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534
Acknowledgements
We are thankful to KIT Germany for provision of necessary training to execute this study in Pakistan and Humboldt foundation for sponsoring the post doc training of the corresponding author and provision of equipment subsidy for smooth execution of experimentation at CEMB Pakistan.
Funding
The research was conducted by in house facilities of CEMB Center of Excellence in Molecular Biology (CEMB), without any grant. However, the student bench fee was provided by Education Commission (HEC) of Pakistan.
Author information
Authors and Affiliations
Contributions
SS performed all the experiments related to the vector confirmation, genetic transformation, compiled data and wrote the initial draft of manuscript. AQR designed the study, constructed recombinant plasmids, provided technical support and supervised the study. SA, AY and MAUK helped in genetic transformation experiments and recorded the data. SS helped in mutant screening and phenotypic analysis. NMA helped in Bioinformatics analysis, FW assisted in designing of protospacer and strategy, MP helped in the provision of chemicals, TH supervised the overall study, read the manuscript critically and presented it in its current form.
Corresponding author
Ethics declarations
Conflict of interest
The authors Sana Shakoor, Abdul Qayyum Rao, Sara Ajmal, Aneela Yasmeen, Muhammad Azmat Ullah Khan, Sahar Sadaqat, Naeem Mahmood Ashraf, Felix Wolter, Michael Pacher and Tayyab Husnain declare that they have no conflicts of interest in submitting or publishing this manuscript entitled “Multiplex Cas9-based excision of CLCuV betasatellite and DNA-A revealed reduction of viral load with asymptomatic cotton plants” in the Journal of Planta.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shakoor, S., Rao, A.Q., Ajmal, S. et al. Multiplex Cas9-based excision of CLCuV betasatellite and DNA-A revealed reduction of viral load with asymptomatic cotton plants. Planta 258, 79 (2023). https://doi.org/10.1007/s00425-023-04233-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04233-w