Abstract
Main conclusion
The outcome of different host–pathogen interactions is influenced by both genetic and epigenetic systems, which determine the response of plants to pathogens and vice versa. This review highlights key molecular mechanisms and conceptual advances involved in epigenetic research and the progress made in epigenetics of wheat-rust interactions.
Abstract
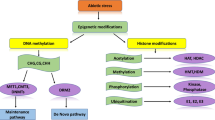
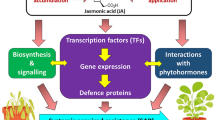
Epigenetics implies the heritable changes in the way of gene expression as a consequence of the modification of DNA bases, histone proteins, and/or non-coding-RNA biogenesis without disturbing the underlying nucleotide sequence. The changes occurring between DNA and its surrounding chromatin without altering its DNA sequence and leading to significant changes in the genome of any organism are called epigenetic changes. Epigenetics has already been used successfully to explain the mechanism of human pathogens and in the identification of pathogen-induced modifications within various host plants. Wheat rusts are one of the most vital fungal diseases throughout the major wheat-growing areas of the world. The epigenome in plant pathogens causing diseases such as wheat rusts is mysterious. The investigations of host and pathogen epigenetics in the wheat rusts system can offer a piece of suitable evidence for elucidation of the molecular basis of host–pathogen interaction. Besides, the information on the epigenetic regulation of the genes involved in resistance or pathogenicity will provide better insights into the complex resistance signaling pathways and could provide answers to certain key questions, such as whether epigenetic regulation of certain genes is imparting resistance to host in response of certain pathogen elicitors or not. In the last few years, there has been an upsurge in research on the host as well as pathogen epigenetics and its outcome in plant-pathogen interactions. This review summarizes the progress made in the areas related to the epigenetic control of host–pathogen interaction with particular emphasis on wheat rusts.


Similar content being viewed by others
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Aina R, Sgorbati S, Santagostino A, Labra M, Ghiani A, Citterio S (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121:472–480
Ali S, Laurie JD, Linning R, Cervantes-Chavez JA, Gaudet D, Bakkeren G (2014) An immunity-triggering effector from the Barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. Plos Pathog 10:e1004223
Allfrey VG, Mirsky AE (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci 51:786–794
Allshire RC, Ekwall K (2015) Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol 7:a018770
Alonso C, Perez R, Bazaga P, Medrano M, Herrera CM (2014) Individual variation in size and fecundity is correlated with differences in global DNA cytosine methylation in the perennial herb Helleborus foetidus (Ranunculaceae). Am J Bot 101:1309–1313
Alonso CP, Erez R, Bazaga P, Herrera CM (2015) Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms. Front Genet 29(6):4
Aramayo R, Selker EU (2013) Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol 5:a017921
Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37:139–143
Aydin M, Taspinar MS, Cakmak ZE, Dumlupinar R, Agar G (2016) Static magnetic field induced epigenetic changes in wheat callus. Bioelectromagnetics 37(7):504–511
Bannister A, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395
Baum S, Reimer-Michalski EM, Jaskiewicz MR et al (2020) Formaldehyde-assisted isolation of regulatory DNA elements from Arabidopsis leaves. Nat Protoc 15:713–733
Beisson J, Sonneborn TM (1965) Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. PNAS 53(2):275–282
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A (2009) An operational definition of epigenetics. Genes Dev 23:781–783
Bhavsar A, Guttman J, Finlay B (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449:827–861
Bird AP (1978) Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol 118:49–60
Bird AP, Southern EM (1978) Use of restriction enzymes to study eukaryotic DNA methylation: the methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol 118:27–47
Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159:266–285
Borrill P, Harrington SA, Uauy C (2019) Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J 97(1):56–72
Boyko A, Kovalchuk I (2011) Genetic and epigenetic effects of plant-pathogen interactions: an evolutionary perspective. Mol Plant 4(6):1014–1023
Brenchley R, Spannagl M, Pfeifer M et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Felsenfeld G (2014) A brief history of epigenetics. CD Allis, ML Caparros, T Jenuwein, D Reinberg (eds.). Additional Perspectives on Epigenetics. Cold Spring Harb Perspect Biol 6: a018200.
Brink RA (1956) A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41:872–889
Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843–851
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109:21.29.1-21.29.9
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6(5):351–360
Chandler VL, Eggleston WB, Dorweiler JE (2000) Paramutation in maize. Plant Mol Biol 43:121–145
Chen ZJ, Tian L (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769:295–307
Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genom 277:589–600
Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S et al (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotech 4:265–270
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD et al (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219
Conrath BGJU, Langenbach CJ, JaskiewiczM R (2015) Priming for enhanced defence. Annu Rev Phytopathol 53:97–119
Cui K, Zhao K (2012) Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol Biol 833:413–419
Dakouri A, McCallum BD, Radovanovic NSC (2013) Mo;ecular and phenotypic characterization of seedlings and adult plant lead rust resistance in a world wheat collection. Mol Breeding 32:663–667
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 16:746–751
De Wit PJG (2007) How plants recognize pathogens and defend themselves. Cell Mol Life Sci 64:2726–2732
Deepak SA, Kottapalli KR, Rakwal R, Oros G, Rangappa KS, Iwahashi H, Agrawal MY, GK, (2007) Real-time PCR: revolutionizing detection and expression analysis of genes. Curr Genom 8(4):234–251
Ding S, Mehrabi R, Koten C, Kang Z, Wei Y, Seong K, Kistler HC, Xu JR (2009) Transducin beta-like gene FTL1 is essential for pathogenesis in Fusarium graminearum. Eukaryot Cell 8(6):867–876
Ding L, Xu H, Yi H, Yang L, Kong Z, Zhang L, Kong Z, Zhang L, Xue S, Jia H, Ma Z (2011) Resistance to Hemi-Biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE 6(4):e19008
Dixon RA (2001) Natural products and plant disease resistance. Nature 411:843–847
Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale SD, Sui Y, Zhang Z, Dou D, Zheng X, Gijzen M, Tyler BM, Wang Y (2011) Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. Plos Pathog 7:2353
Down TA, Rakyan VK, Turner DJ, Flicek P, Li H et al (2008) A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26:779–785
Dubey A, Jeon J (2017) Epigenetic regulation of development and pathogenesis in fungal plant pathogens. Mol Plant Patho 18(6):887–898
Dubey H, Kiran K, Jaswal R et al (2019) Discovery and profiling of small RNAs from Puccinia triticina by deep sequencing and identification of their potential targets in wheat. Funct Integr Genom 19:391
Eisenman HC, Casadevall A (2012) Synthesis and assembly of fungal melanin. Appl Microbiol Biot 93:931–940
Eversmeyer MG, Browder LE (1974) Effect of leaf and stem rust in 1973 Kansas wheat yields. Plant Dis Rep 58:469–471
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331–368
Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Nati Acad Sci USA 93:8449–8454
Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC et al (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defence. Annu Rev Plant Biol 64:839–863
Fu DL, Uauy C, Distelfeld A et al (2009a) A kinase-START Gene confers temperature-dependent resistance to wheat stripe rust. Science 323(5919):1357–1360
Fu SJ, Wang H, Feng LN, Sun Y, Yang WX, Liu DQ (2009b) Analysis of methylation-sensitive amplified polymorphism in wheat genome under the wheat leaf rust stress. Yi Chuan 31(3):297–304
Gacek A, Strauss J (2012) The chromatin code of fungal secondary metabolite gene clusters. Appl Microbiol Biot 95:1389–1404
Garnica DP, Upadhyaya NM, Dodds PN, Rathjen JP (2013) Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS ONE 8(6):e67150
Gijzen M, Ishmael C, Shrestha SD (2014) Epigenetic control of effectors in plant pathogens. Front Plant Sci 5:638
Gilroy EM, Breen S, Whisson SC et al (2011) Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol 191:763–776
Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD (2007) FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 17(6):877–885
Gohlke J, Scholz CJ, Kneitz S, Weber D, Fuchs J, Hedrich R, Deeken R (2013) DNA methylation mediated control of gene expression is critical for development of crown gall tumors. Plos Genet 9(2):e1003267
Gomez-Diaz E, Jorda M, Peinado MA, Rivero A (2012) Epigenetics of host–pathogen interactions: The road ahead and the road behind. Plos Pathog 8(11):e1003007
Grunstein M, Gasser SM (2013) Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5:a017491–a017491
Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A (2011) Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 6(4):468–481
Hamon MA, Cossart P (2008) Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4:100–109
Henikoff S (2005) Histone modifications: combinatorial complexity or cumulative simplicity? PNAS 102(15):5308–5309
Herrera CM, Bazaga P (2010) Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol 187(3):867–876
Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22:115–122
Holliday R, Pugh JE (1975) DNA modification mechanisms and gene activity during development. Science 187:226–232
Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16(2):71–84
Hurd H (2009) Evolutionary drivers of parasite-induced changes in insect life-history traits: from theory to underlying mechanisms. In: Joanne PW (ed) Advances in parasitology. Academic Press, Cambridge, pp 85–110
Hussain M, Iqbal MA, Till BJ, Rahman M (2018) Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS ONE 13:e0201918
Ivashuta S, Naumkina M, Gau M et al (2002) Genotype-dependent transcriptional activation of novel repetitive elements during cold acclimation of alfalfa (Medicago sativa). Plant J 31(5):615–627
Jablonka E, Lamb MJ (2010) Transgenerational epigenetic inheritance. In: Pigliucci M, Müller GB (eds) Evolution—the extended synthesis. MIT Press, Cambridge, pp 137–174
Jain N, Rani S, Sharma C et al (2020) Large-scale stage-specific regulation of gene expression during host-pathogen interactions in CSP44 bread wheat carrying APR gene Lr48. Func Plant Biol 47(3):203–225
Jeon J, Kwon S, Lee YH (2014) Histone acetylation in fungal pathogens of plants. Plant Pathol J 30:1–9
Jeon J, Choi J, Lee GW, Park SY, Huh A, Dean RA, Lee YH (2015) Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe Oryzae. Sci Rep 5:8567
Jończyk M, Sobkowiak A, Trzcinska-Danielewicz J et al (2021) Chromatin-level differences elucidate potential determinants of contrasting levels of cold sensitivity in maize lines. Plant Mol Biol Rep 39:335–350
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jorda M, Rodriguez J, Frigola J, Peinado MA (2009) Analysis of DNA methylation by amplification of intermethylated sites (AIMS). In: Tost J (ed) DNA methylation: methods and protocols, 2nd edn. Humana Press, Totowa, pp 107–116
Kang S, Lebrun MH, Farrall L, Valent B (2001) Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol Plant-Microbe Interact 14:671–674
Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I (2010) Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol 153:1859–1870
Kaur J, Fellers J, Adholeya A, Velivelli SL, El-Mounadi K, Nersesian N, Clemente T, Shah D (2017) Expression of apoplast-targeted plant defensin MtDef4.2 confers resistance to leaf rust pathogen Puccinia triticina but does not affect mycorrhizal symbiosis in transgenic wheat. Transgenic Res 26(1):37–49
Kelsey A, Maher MB, Kajala K et al (2018) Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30(1):15–36
Kong L, Liu Y, Wang X, Chang C (2020) Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. IJMS 21(1480):1–15
Kumar S, Singh AK, Mohapatra T (2017) Epigenetics: history, present status and future perspective. Indian J Genet Plant Breed 77:445–463
Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4:694–699
Laudencia-Chingcuanco D, Ganeshan S, You F, Fowler B, Chibbar R, Anderson O (2011) Genome-wide gene expression analysis supports a developmental model of low temperature tolerance gene regulation in wheat (Triticum aestivum L.). BMC Genomics 12:299
Lee EJ, Pei L, Srivastava G, Joshi T, Kushwaha G, Choi JH et al (2011) Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucl Acids Res 39(19):e127
Lefevre T, Lebarbenchon C, Gauthier-Clerc M, Misse´ D, Poulin R, et al (2009) The ecological significance of manipulative parasites. Trends Ecol Evol 24:41–48
Li LC, Qin GJ, Tsuge T et al (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179:751–764
Li Y, Wang C, Liu W, Wang G, Zhensheng Kang H, Kistler C, Jin-Rong Xu (2011) The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum. Mol Plant-Microbe Interact 24(4):487–496
Li Z, Wang M, Lin K et al (2019) The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20:139
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322
Liu X, Huang J, Parameswaran S et al (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151(3):1401–1411
Liu S, Li F, Kong L, Sun Y, Qin L, Chen S, Cui H, Huang Y, Xia G (2015) Genetic and epigenetic changes in somatic hybrid introgression lines between wheat and tall wheatgrass. Genetics 199:1035–1045
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819(2):129–136
Lyon MF (1961) Gene action in the X-chromosome of the mouse. Nature 190:372–373
Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Nikolai M, Adamski LB, McIntosh R, Wulff BBH, Berry S, Lagudah E, Uauy C (2018) BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants 4:662–668
Mason G, Noris E, Lanteri S, Acquadro A, Accotto GP, Portis E (2008) Potentiality of methylation-sensitive amplification polymorphism (MSAP) in identifying genes involved in tomato response to tomato yellow leaf curl Sardinia virus. Plant Mol Biol Rep 26:156–173
Mayer KFX, Taudien S, Martis M et al (2009) Gene content and virtual gene order of barley chromosome1H. Plant Physiol 151:496–505
Mayer KFX, Martis M, Hedley PE et al (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23:1249–1263
McClintock B (1965) The control of gene action in maize. Brookhaven Symp Biol 18:162–184
Mendes MA, Petrella R, Cucinotta M, Vignati E, Gatti S, Pinto SC, Bird DC, Gregis V, Dickinson H, Tucker MR, Colombo L (2020) The RNA-dependent DNA methylation pathway is required to restrict SPOROCYTELESS/NOZZLE expression to specify a single female germ cell precursor in Arabidopsis. Development. https://doi.org/10.1242/dev.194274
Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51:245–266
Mohammad P, Golicz AA, Bhalla PL, Singh MB (2020) Global role of crop genomics in the face of climate change. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00922
Molinier J, Ries G, Zipfel C, Hohn B (2006) Transgenerational memory of stress in plants. Nature 442:1046–1049
Mueth NA, Ramachandran SR, Hulbert SH (2015) Small RNAs from the wheat stripe rust fungus (Puccinia striiformis f.sp. tritici). BMC Genomics 16:718
Na R, Yu D, Chapman BP, Zhang Y, Kuflu K, Austin R et al (2014) Genome re-sequencing and functional analysis places the Phytophthora sojae avirulence genes Avr1c and Avr1a in a tandem repeat at a single locus. PLoS ONE 9:e89738
Nigam D, Kavita P, Tripathi RK, Ranjan A, Goel R, Asif M, Shukla A, Singh G, Rana D, Sawant SV (2014) Transcriptome dynamics during fibre development in contrasting genotypes of Gossypium hirsutum L. Plant Biotechnol J 12:204–218
Nutzmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA (2011) Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. PNAS 108(14):282–287
Ohno S, Kaplan WD, Kinosita R (1959) Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp Cell Res 18:415–418
Panwar V, McCallum B, Bakkeren G (2013) Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol Biol 81(6):595–608
Panwar V, Jordan M, McCallum B, Bakkeren G (2018) Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol J 16(5):1013–1023
Paschos K, Allday MJ (2010) Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol 18:439–447
Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, Linder-Basso D, Plachetka A, Shanower G, Tolstorukov MY, Luquette LJ, Xi R, Jung YL, Park RW, Bishop EP, Canfield TK, Sandstrom R, Thurman RE, MacAlpine DM, John A., Kellis SM, Elgin SCR, Kuroda MI, Pirrotta V, Karpen GH, Park PJ (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471(7339):480–485. https://doi.org/10.1038/nature09725
Picelli S, Bjorklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R (2014) Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24(12):2033–2040
Pieterse CM, Leon-Reyes A, Van Der Ent S, Van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5:308–316
Platt A, Gugger P, Sork VL (2015) Genome-wide signature of local adaptation linked to variable CpG methylation in oak populations. Mol Ecol 24:3823–3830
Prasad P, Savadi S, Bhardwaj SC, Gangwar OP, Kumar S (2019) Rust pathogen effectors: perspectives in resistance breeding. Planta 250:1–22
Qi T, Wang J, Sun Q, Day B, Guo J, Ma Q (2017) TaARPC3, contributes to wheat resistance against the stripe rust fungus. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01245
Qiu T, Dong YZ, Yu XM, Zhao N, Yang YF (2017) Analysis of allopolyploidy-induced rapid genetic and epigenetic changes and their relationship in wheat. Genet Mol Res. https://doi.org/10.4238/gmr16029303
Qutob D, Tedman-Jones J, Dong S, Kuflu K, Pham H, Wang Y, Dou D, Kale SD, Arredondo FD, Gijzen TBM, M, (2009) Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS ONE 4:5066
Qutob D, Chapman BP, Gijzen M (2013) Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat Commun 4:1349
Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F (2006) RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441:469–474
Raut VV, Shashibhal MP, Jayashree KS (2011) Histone octamer trans-transfer: a signature mechanism of ATP-dependent chromatin remodelling unravelled in wheat nuclear extract. Ann Bot 108(7):1235–1246
Rebollo R, Horard B, Horard B, Vieira C (2010) Jumping genes and epigenetics: towards new species. Gene 454:1–7
Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J (2010) Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol 76:1376–1386
Richards CL, Alonso C, Becker C, Bossdorf O, Bucher E, Colome-Tatche M, Walter Durka D, Engelhardt J, Gaspar B, GogoL-Doring A, Grosse I, van Gurp TP, Heer K, Kronholm I, Lampei C, Latzel V, Mirouze M, Opgenoorth L, Paun O, Prohaska SJ, Rensing SA, Stadler PF, Trucchi E, Ullrich K, Verhoeven KJF (2017) Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecol Lett 20:1576–1590
Riggs AD (1975) X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14:9–25
Rizzo JM, Sinha S (2014) Analyzing the global chromatin structure of keratinocytes by MNase-seq. Methods Mol Biol 1195:49–59
Robertson MH, Schrey AW, Shayter A, Moss CJ, Richards CL (2017) Genetic and epigenetic variation in Spartina alterniflora following the Deepwater Horizon oil spill. Evol Appl. https://doi.org/10.1111/eva.12482
Rovenich H, Boshoven JC, Thomma BPHJ (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol 20:96–103
Saripalli G, Sharma C, Gautam T, Singh K, Jain N, Prasad P et al (2020) Complex relationship between DNA methylation and gene expression due to Lr28 in wheat-leaf rust pathosystem. Mol Biol Rep 47(2):1339–1360
Savadi S, Prasad P, Kashyap PL, Bhardwaj SC (2018) Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol 67:771–791
Scharf DH, Heinekamp T, Brakhage AA (2014) Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog 10:e1003859
Shan W, Cao M, Leung D, Tyler BM (2004) The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps 1b. Mole Plant Microbe Interact 17:394–403
Shapiro JA (2010) Mobile DNA and evolution in the 21st century. Mob DNA 1:4
Sharma I, Prasad P, Bhardwaj SC (2017) Recent molecular technologies for tackling wheat diseases. In: Langridge P (ed) Achieving sustainable cultivation of wheat, vol 1. Burleigh Dodds Science Publishing Limited, Cambridge, pp 385–416
Sharma C, Kumar S, Saripalli G, Jain N, Raghuvanshi S, Sharma JB, Prabhu KV, Sharma PK, Balyan HS, Gupta PK (2019) H3K4/K9 acetylation and Lr28-mediated expression of six leaf rust responsive genes in wheat (Triticum aestivum). Mol Genet Genom 294(1):227–241
Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K et al (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19:1723–1737
Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. NatRev Genet 8:272–285
Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ (2012) Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. Plos Pathog 8:e1002514
Spiker S, Murraytt MG, Thompson WF (1983) DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc Natl Acad Sci USA 80:815–819
Stam M, Belele C, Dorweiler JE, Chandler VL (2002) Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev 16:1906–1918
Stamatoyannopoulos JA, Kellis M, Elgin SC, Kuroda MI, Pirrotta V, Karpen GH, Park PJ (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471(7339):480–485
Stedman E, Stedman E (1950) Cell specificity of histones. Nature 166:780–781
Stergiopoulos I, De Wit PJGM (2009) Fungal effector proteins. Annu Rev Phytopathol 47:233–263
Steuernagel B, Periyannan S, Hernández-Pinzón I, Witek K, Yu RMN, Hatta G et al (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34:652–655
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765):41–45
Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152(1–2):352–364
Taunton J, Hassig CA, Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408–411
Thind AK, Wicker T, Simkova H, Fossati D, Moullet O, Brabant C, Vrana J, Doležel J, Krattinger SG (2017) Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat Biotechnol 35:793–796
Thomas M, Pingault L, Poulet A et al (2014) Evolutionary history of Methyltransferase 1 genes in hexaploid wheat. BMC Genomics 15:922
Trucchi E, Mazzarella AB, Gilfillan GD, Romero MT, Paun O (2016) BsRADseq screening DNA methylation in natural populations of non-model species. Mol Ecol 25:1697–1713
Tsompana M, Buck MJ (2014) Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7:33
Van Der Hoorn RAL, Wit PJGMD, Joosten MHAJ (2002) Balancing selection favors guarding resistance proteins. Trends Plant Sci 7:67–71
Van Gurp TP, Wagemaker NCAM, Wouters B, Vergeer P, Ouborg JNJ, Verhoeven KJF (2016) epiGBS: reference-free reduced representation bisulfite sequencing. Nat Meth 13:322–324
Venetsky A, Levy-Zamir A, Khasdan V, Domb K, Kashkush K (2015) Structure and extent of DNA methylation-based epigenetic variation in wild emmer wheat (T. turgidum ssp. dicoccoides) populations. BMC Plant Biol 15:200
Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A (2010) Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 185:1108–1118
Vleeshouwers VG, Oliver RP (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27(3):196–206
Wada Y, Miyamoto K, Kusano T, Sano H (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genomics 271:658–666
Wang Y, Jiang R, Wong WH (2016) Modeling the causal regulatory network by integrating chromatin accessibility and transcriptome data. Natl Sci Rev 3:240–251
Waterborg JH (2011) Plant histone acetylation: in the beginning. Biochim Biophys Acta 1809:353–359
Xu L, Huang H (2014) Genetic and epigenetic controls of plant regeneration. Curr Top Dev Biol 108:1–33
Yin C, Jurgenson JE, Hulbert SH (2011) Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol Plant Microbe Interact 24(5):554–561
Yong Z, Hui LZ, Cheng L, Jun YZ, Jun DK, Hua PJ, Ping ZJ, Rong LG, Xiang TZ, Long RZ (2008) Analysis of DNA methylation variation in wheat genetic background after alien chromatin introduction based on methylation-sensitive amplification polymorphism. Science 53(1):58–69
Zhang W, Jiang J (2015) Genome-wide mapping of DNase I hypersensitive sites in plants. Methods Mol Biol 1284:71–89
Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007) Whole-genome analysis of histone H3 Lysine 27 trimethylation in Arabidopsis. Plos Biol 5(5):e129
Zhang H, Chen X, Wang C, Xu Z, Wang Y, Liu X, Kang Z, Ji W (2013) Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol Biol Rep 40(11):6245–6253
Zhang C, Huang L, Zhang H et al (2019) An ancestral NB-LRR with duplicated 3′UTRs confers stripe rust resistance in wheat and barley. Nat Commun 10:4023
Zhu QH, Shan WX, Ayliffe MA, Wang MB (2016) Epigenetic mechanisms: an immerging player in plant-microbe interactions. Mol Plant-Microbe Interact 29(3):187–196
Acknowledgements
We sincerely thank Director, Indian Council of Agricultural Research (ICAR)-Indian Institute of Wheat and Barley Research (IIWBR), Karnal, India for financial and liberal support.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No conflict of interest among the authors.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shilpa, Thakur, R.K., Prasad, P. et al. Epigenetics of wheat–rust interaction: an update. Planta 255, 50 (2022). https://doi.org/10.1007/s00425-022-03829-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03829-y