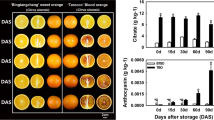
Abstract
Main conclusion
4-coumarate-CoA ligase (VIT_02s0109g00250) and copper amine oxidase (VIT_17s0000g09100) played essential roles in contributing to the total soluble solid and total anthocyanin variations induced by bud sport in grape berries.
Abstract
Taste and color, which are important organoleptic qualities of grape berry, undergo rapid and substantial changes during development and ripening. In this study, we used two cultivars ‘Summer Black’ and its bud sport ‘Nantaihutezao’ to explore and identify differentially expressed genes associated with total soluble solid and anthocyanin during developmental stages using RNA-Seq. Overall, substantial differences in expression were observed across berry development between the two cultivars. 5388 genes were detected by weighted gene co-expression network analysis (WGCNA) associated with the total soluble solid (TSS) and anthocyanin contents variations. Several of these genes were significantly enriched in the phenylalanine metabolism pathway; two hub genes 4-coumarate-CoA ligase (VIT_02s0109g00250) and copper amine oxidase (VIT_17s0000g09100) played the most essential roles in relating to the total soluble solid and total anthocyanin variations induced by bud sport through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and co-expression network analysis. These findings provide insights into the molecular mechanism responsible for the bud sport phenotype.







Similar content being viewed by others
Abbreviations
- WGCNA:
-
Weighted gene co-expression network analysis
- TSS:
-
Total soluble solid
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Azuma A, Kobayashi S, Goto-Yamamoto N, Shiraishi M, Mitani N, Yakushiji H, Koshita Y (2009) Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus. Plant Sci 176(4):470–478. https://doi.org/10.1016/j.plantsci.2008.12.015
Azuma A, Yakushiji H, Koshita Y, Kobayashi S (2012) Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236(4):1067–1080. https://doi.org/10.1007/s00425-012-1650-x
Balic I, Vizoso P, Nilo-Poyanco R, Sanhueza D, Olmedo P, Sepulveda P, Arriagada C, Defilippi BG, Meneses C, Campos-Vargas R (2018) Transcriptome analysis during ripening of table grape berry cv. Thompson seedless. PLoS ONE 13(1):e190087. https://doi.org/10.1371/journal.pone.0190087
Berli FJ, Moreno D, Piccoli P, Hespanhol-Viana L, Silva MF, Bressan-Smith R, Cavagnaro JB, Bottini R (2010) Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ 33(1):1–10. https://doi.org/10.1111/j.1365-3040.2009.02044.x
Blanco-Ulate B, Amrine KCH, Collins TS, Rivero RM, Vicente AR, Morales-Cruz A, Doyle CL, Ye ZR, Allen G, Heymann H, Ebeler SE, Cantu D (2015) Developmental and metabolic plasticity of white-skinned grape berries in rresponse to Botrytis cinerea during noble rot. Plant Physiol 169(4):2422–2443. https://doi.org/10.1104/pp.15.00852
Carbonell-Bejerano P, Maria ES, Torres-Perez R, Royo C, Lijavetzky D, Bravo G, Aguirreolea J, Sanchez-Diaz M, Antolin MC, Martinez-Zapater JM (2013) Thermotolerance responses in ripening berries of Vitis vinifera L. cv Muscat Hamburg. Plant Cell Physiol 54(7):1200–1216. https://doi.org/10.1093/pcp/pct071
Castellarin SD, Matthews MA, Di Gaspero G, Gambetta GA (2007) Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 227(1):101–112. https://doi.org/10.1007/s00425-007-0598-8
Castellarin SD, Gambetta GA, Wada H, Shackel KA, Matthews MA (2011) Fruit ripening in Vitis vinifera: spatiotemporal relationships among turgor, sugar accumulation, and anthocyanin biosynthesis. J Exp Bot 62(12):4345–4354. https://doi.org/10.1093/jxb/err150
Chen H, Boutros PC (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. https://doi.org/10.1186/1471-2105-12-35
Cohen SD, Tarara JM, Gambetta GA, Matthews MA, Kennedy JA (2012) Impact of diurnal temperature variation on grape berry development, proanthocyanidin accumulation, and the expression of flavonoid pathway genes. J Exp Bot 63(7):2655–2665. https://doi.org/10.1093/jxb/err449
Degu A, Hochberg U, Sikron N, Venturini L, Buson G, Ghan R, Plaschkes I, Batushansky A, Chalifa-Caspi V, Mattivi F, Delledonne M, Pezzotti M, Rachmilevitch S, Cramer GR, Fait A (2014) Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol 14:188. https://doi.org/10.1186/s12870-014-0188-4
Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, Pais MS (2011) Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol 11:149. https://doi.org/10.1186/1471-2229-11-149
Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S (2006) Integrating genetic and network analysis to characterize genes related to mouse weight. Plos Genet 2(8):1182–1192. https://doi.org/10.1371/journal.pgen.0020130
Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8:187. https://doi.org/10.1186/1471-2164-8-187
Guo DL, Xi FF, Yu YH, Zhang XY, Zhang GH, Zhong GY (2016) Comparative RNA-Seq profiling of berry development between table grape ‘Kyoho’ and its early-ripening mutant ‘Fengzao.’ BMC Genomics 17:795. https://doi.org/10.1186/s12864-016-3051-1
Jaillon O, Aury JM, The French–Italian Public Consortium for Grapevine Genome Characterization et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449(7161):463–467. https://doi.org/10.1038/nature06148
Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka A (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci 167(2):247–252. https://doi.org/10.1016/j.plantsci.2004.03.021
Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M (2006) Expression of the flavonoid 3’-hydroxylase and flavonoid 3’,5’-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci 170(1):61–69. https://doi.org/10.1016/j.plantsci.2005.07.025
Ju YL, Liu M, Tu TY, Zhao XF, Yue XF, Zhang JX, Fang YL, Meng JF (2018) Effect of regulated deficit irrigation on fatty acids and their derived volatiles in ‘Cabernet Sauvignon’ grapes and wines of Ningxia, China. Food Chem 245:667–675. https://doi.org/10.1016/j.foodchem.2017.10.018
Kayesh E, Shangguan LF, Korir NK, Sun X, Bilkish N, Zhang YP, Han J, Song CN, Cheng ZM, Fang JG (2013) Fruit skin color and the role of anthocyanin. Acta Physiol Plant 35(10):2879–2890. https://doi.org/10.1007/s11738-013-1332-8
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. https://doi.org/10.1186/Gb-2013-14-4-R36
Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose : flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160(3):543–550. https://doi.org/10.1016/S0168-9452(00)00425-8
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304(5673):982. https://doi.org/10.1126/science.1095011
Kuhn N, Guan L, Dai ZW, Wu BH, Lauvergeat V, Gomes E, Li SH, Godoy F, Arce-Johnson P, Delrot S (2014) Berry ripening: recently heard through the grapevine. J Exp Bot 65(16):4543–4559. https://doi.org/10.1093/jxb/ert395
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Leng F, Lin Q, Wu D, Wang SP, Wang DL, Sun CD (2016) Comparative transcriptomic analysis of grape berry in response to root restriction during developmental stages. Molecules 21:1431. https://doi.org/10.3390/Molecules21111431
Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, Fenoll J, Hellin P, Oliveros JC, Martinez-Zapater JM (2012) Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PLoS ONE 7(6):e3954. https://doi.org/10.1371/journal.pone.0039547
Liu XT, Zhai R, Feng WT, Zhang SW, Wang ZG, Qiu ZH, Zhang JK, Ma FW, Xu LF (2014) Proteomic analysis of ‘Zaosu’ pear (Pyrus bretschneideri Rehd.) and its early-maturing bud sport. Plant Sci 224:120–135. https://doi.org/10.1016/j.plantsci.2014.04.012
Lorrain B, Ky I, Pasquier G, Jourdes M, Dubrana LG, Geny L, Rey P, Doneche B, Teissedre PL (2012) Effect of Esca disease on the phenolic and sensory attributes of Cabernet Sauvignon grapes, musts and wines. Aust J Grape Wine Res 18(1):64–72. https://doi.org/10.1111/j.1755-0238.2011.00172.x
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/S13059-014-0550-8
Martinez-Luscher J, Torres N, Hilbert G, Richard T, Sanchez-Diaz M, Delrot S, Aguirreolea J, Pascual I, Gomes E (2014) Ultraviolet-B radiation modifies the quantitative and qualitative profile of flavonoids and amino acids in grape berries. Phytochemistry 102:106–114. https://doi.org/10.1016/j.phytochem.2014.03.014
Mori K, Saito H, Goto-Yamamoto N, Kitayama M, Kobayashi S, Sugaya S, Gemma H, Hashizume K (2005) Effects of abscisic acid treatment and night temperatures on anthocyanin composition in Pinot noir grapes. Vitis 44(4):161–165. https://doi.org/10.1016/j.pnpbp.2004.11.023
Osorio S, Scossa F, Fernie AR (2013) Molecular regulation of fruit ripening. Front Plant Sci 4:198. https://doi.org/10.3389/Fpls.2013.00198
Pang YJ, Li HY, Zhu XH, Gao FM, Yin YM, Jia HJ (2017) Genetic identification of bud sport strain ‘11-06-25’ from ‘Sanbenti’ grape. J Zhejiang Univ Agric Life Sci 43(1):73–80. https://doi.org/10.3785/j.issn.1008-9209.2016.06.201
Pilati S, Perazzolli M, Malossini A, Cestaro A, Dematte L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C (2007) Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genomics 8:428. https://doi.org/10.1186/1471-2164-8-428
Pillet J, Egert A, Pieri P, Lecourieux F, Kappel C, Charon J, Gomes E, Keller F, Delrot S, Lecourieux D (2012) VvGOLS1 and VvHsfA2 are Involved in the heat stress responses in grapevine berries. Plant Cell Physiol 53(10):1776–1792. https://doi.org/10.1093/pcp/pcs121
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Sweetman C, Wong DCJ, Ford CM, Drew DP (2012) Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genomics 13:691. https://doi.org/10.1186/1471-2164-13-691
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. https://doi.org/10.1038/nbt.1621
Vega A, Gutierrez RA, Pena-Neira A, Cramer GR, Arce-Johnson P (2011) Compatible GLRaV-3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera. Plant Mol Biol 77(3):261–274. https://doi.org/10.1007/s11103-011-9807-8
Walker AR, Lee E, Robinson SP (2006) Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Mol Biol 62(4–5):623–635. https://doi.org/10.1007/s11103-006-9043-9
Wang LG, Wang SQ, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28(16):2184–2185. https://doi.org/10.1093/bioinformatics/bts356
Wu JM, Mao XZ, Cai T, Luo JC, Wei LP (2006) KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 34:W720–W724. https://doi.org/10.1093/nar/gkl167
Xi FF, Guo LL, Yu YH, Wang Y, Li Q, Zhao HL, Zhang GH, Guo DL (2017) Comparison of reactive oxygen species metabolism during grape berry development between ‘Kyoho’ and its early ripening bud mutant ‘Fengzao.’ Plant Physiol Biochem 118:634–642. https://doi.org/10.1016/j.plaphy.2017.08.007
Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. https://doi.org/10.1093/nar/gkr483
Xu WR, Li RM, Zhang NB, Ma FL, Jiao YT, Wang ZP (2014) Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol Biol 86(4–5):527–541. https://doi.org/10.1007/s11103-014-0245-2
Xu YS, Jiang N, Zhang YG, Wang M, Ren JP, Tao JM (2017) A SNP in the promoter region of the VvmybA1 gene is responsible for differences in grape berry color between two related bud sports of grape. Plant Growth Regul 82(3):457–465. https://doi.org/10.1007/s10725-017-0272-5
Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152(4):1787–1795. https://doi.org/10.1104/pp.109.149716
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:17. https://doi.org/10.2202/1544-6115.1128
Zhang YJ, Wang XJ, Wu JX, Chen SY, Chen H, Chai LJ, Yi HL (2014) Comparative transcriptome analyses between a spontaneous late-ripening sweet orange mutant and its wild type suggest the functions of ABA, sucrose and JA during citrus fruit ripening. PLoS ONE 9(12):e116056. https://doi.org/10.1371/journal.pone.0116056
Zhang KK, Liu ZJ, Guan L, Zheng T, Jiu ST, Zhu XD, Jia HF, Fang JG (2018) Changes of anthocyanin component biosynthesis in ‘Summer Black’ grape berries after the red flesh mutation occurred. J Agric Food Chem 66(35):9209–9218. https://doi.org/10.1021/acs.jafc.8b02150
Zhao Y, Zhao X, Zhao S, Han N (2015) A novel bud sport fromthe ‘Benitaka’ table grape cultivar (Vitis vinifera L.) improves sugar and anthocyanin accumulation at the berry ripening stage. S Afr J Bot 97:111–116. https://doi.org/10.1016/j.sajb.2014.12.011
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20190899); the doctor for entrepreneurship and innovation program of Jiangsu Province; the Yangzhou City's Green and Golden Phoenix Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2020_3543_MOESM1_ESM.jpg
Supplementary file1 Suppl. Fig. S1 Real-time quantitative PCR validation of the candidate genes. Genes differentially expressed between ‘Summer Black’ (black bar chart) and its bud sport ‘Nantaihutezao’ (red bar chart). a S5. b S6. c S7. d S8. e S9. f S10 (JPG 233 KB)
425_2020_3543_MOESM15_ESM.xlsx
Supplementary file15 Suppl. File S14 Weighted gene co-expression network analysis of the genes was positively correlated with TSS and total anthocyanin (XLSX 23 KB)
425_2020_3543_MOESM16_ESM.xlsx
Supplementary file16 Suppl. File S15 Weighted gene co-expression network analysis (WGCNA) of the genes was negatively correlated with TSS and total anthocyanin (XLSX 86 KB)
425_2020_3543_MOESM17_ESM.xlsx
Supplementary file17 Suppl. File S16 KEGG pathway enrichment analyses of genes was positively correlated with TSS and total anthocyanin (XLSX 9 KB)
425_2020_3543_MOESM18_ESM.xlsx
Supplementary file18 Suppl. File S17 KEGG pathway enrichment analyses of genes was negatively correlated with TSS and total anthocyanin (XLSX 8 KB)
425_2020_3543_MOESM19_ESM.xlsx
Supplementary file19 Suppl. File S18 The genes involving in the phenylalanine metabolism pathway were employed to construct gene correlation networks (XLSX 10 KB)
Rights and permissions
About this article
Cite this article
Leng, F., Ye, Y., Zhu, X. et al. Comparative transcriptomic analysis between ‘Summer Black’ and its bud sport ‘Nantaihutezao’ during developmental stages. Planta 253, 23 (2021). https://doi.org/10.1007/s00425-020-03543-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03543-7