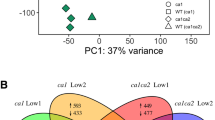
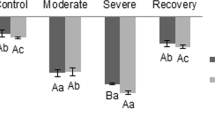
Abstract
Main conclusion
β-carbonic anhydrases, which function in regulating plant growth, C/N status, and stomata number, showed functional redundancy and divergence in Physcomitrella patens.
Abstract
Carbonic anhydrases (CAs) catalyze the interconversion of CO2 and HCO3−. Plants have three evolutionarily unrelated CA families: α-, β-, and γ-CAs. βCAs are abundant in plants and are involved in CO2 assimilation, stress responses, and stomata formation. Recent studies of βCAs have mainly examined C3 or C4 plants, whereas their functions in non-vascular plants are mostly unknown. In this study, phylogenetic analysis revealed that the evolution of βCAs were conserved between subaerial green algae and bryophytes after terrestrialization event, and βCAs from some cyanobacteria might begin evolving for the adaptation of terrestrial environment/habitat. In addition, we investigated the physiological roles of βCAs in the basal land plant Physcomitrella patens. High PpβCA expression levels in different tissues suggest that PpβCAs play important roles in development in P. patens. Plants treated with 1–10 mM NaHCO3 had higher fresh and dry weight, PpβCA expression, total CA activity, and photosynthetic yield (Fv/Fm) compared with water-treated plants. However, treatment with 10 mM NaHCO3 influenced the C/N status. Further study of six Ppβca single-gene mutants revealed that PpβCAs have functional redundancy and divergence in regulating the C/N ratio of plants and stomatal formation. This study provides new insight into the physiological roles of βCAs in basal land plants.







Similar content being viewed by others
Data availability
The plasmids, plant materials and all data used in this study will be freely available once our manuscript is accepted.
Abbreviations
- CA:
-
Carbonic anhydrase
- Pp :
-
Physcomitrella patens
- SV:
-
Splice variant
- WT:
-
Wild-type
References
Alber BE, Ferry JG (1994) A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc Natl Acad Sci USA 91:6909–6913
Badger M (2003) The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosyn Res 77:83e94
Brinkman R, Margaria R, Meldrum N, Roughton F (1932) The CO2 catalyst present in blood. J Physiol 75:3–4
Chater C, Caine RS, Tomek M, Wallace S, Kamisugi Y, Cuming AC et al (2016) Origin and function of stomata in the moss Physcomitrella patens. Nature Plants 2(12):16179. https://doi.org/10.1038/nplants.2016.179
Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T et al (2019) Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179(5):1057–1067
Clayton H, Saladié M, Rolland V, Sharwood R, Macfarlane T, Ludwig M (2017) Loss of the chloroplast transit peptide from an ancestral C3 carbonic anhydrase is associated with C4 evolution in the grass genus Neurachne. Plant Physiol 173(3):1648–1658
Collins RM, Afzal M, Ward DA, Prescott MC, Sait SM, Rees HH, Tomsett AB (2010) Differential proteomic analysis of Arabidopsis thaliana genotypes exhibiting resistance or susceptibility to the insect herbivore Plutella xylostella. PLoS ONE 5:e10103
DiMario RJ, Quebedeaux JC, Longstreth DJ, Dassanayake M, Hartman MM, Moroney JV (2016) The cytoplasmic carbonic anhydrases βCA2 and βCA4 are required for optimal plant growth at low CO2. Plant Physiol 171(1):280–293
DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV (2017) Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol Plant 10(1):30–46
Dąbrowska-Bronk J, Komar DN, Rusaczonek A, Kozłowska-Makulska A, Szechyńska-Hebda M, Karpiński S (2016) β-carbonic anhydrases and carbonic ions uptake positively influence Arabidopsis photosynthesis, oxidative stress tolerance and growth in light dependent manner. J Plant physiol 203:44–54
Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513:246–250
Fabre N, Reiter IM, Becuwe-Linka N, Genty B, Rumeau D (2007) Characterization and expression analysis of genes encoding α and β carbonic anhydrases in Arabidopsis. Plant Cell Environ 30(5):617–629
Ferreira FJ, Guo C, Coleman JR (2008) Reduction of plastid-localized carbonic anhydrase activity results in reduced Arabidopsis seedling survivorship. Plant Physiol 147(2):585–594
Floryszak-Wieczorek J, Arasimowicz-Jelonek M (2017) The multifunctional face of plant carbonic anhydrase. Plant Physiol Biochem 112:362–368
Foyer CH, Noctor G, Verrier P (2018) Photosynthetic carbon-nitrogen interactions: modelling inter-pathway control and signalling. Annu Plant Rev Online. https://doi.org/10.1002/9781119312994.apr0227
Goodstein D, Batra S, Carlson J, Hayes R, Phillips J, Shu S et al (2014) Phytozome comparative plant genomics portal. Lawrence Berkeley National Laboratory. LBNL Report #: LBNL-7048E. Retrieved from https://escholarship.org/uc/item/22k9d6k9
Hewett-Emmett D, Tashian RE (1996) Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol Phylogenet Evol 5(1):50–77
Hilvo M, Baranauskiene L, Salzano AM, Scaloni A, Matulis D, Innocenti A et al (2008) Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J Biol Chem 283(41):27799–27809
Horton P, Park K, Obayashi T, Fujita N, Harada H, Adamscollier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(Web Server):W585–W587
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Hu H, Boissondernier A, Israelssonnordstrom M, Bohmer M, Xue S, Ries A et al (2010) Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat Cell Biol 12(1):87–93
Jiang CY, Tholen D, Xu JM, Xin CP, Zhang H, Zhu XG, Zhao YX (2014) Increased expression of mitochondria-localized carbonic anhydrase activity resulted in an increased biomass accumulation in Arabidopsis thaliana. J Plant Biol 57:366–374
Joseph P, Turtaut F, Ouahrani-Bettache S, Montero JL, Nishimori I, Minakuchi T et al (2010) Cloning, characterization, and inhibition studies of a β-carbonic anhydrase from Brucella suis. J Med Chem 53(5):2277–2285
Jung HW, Lim CW, Lee SC, Choi HW, Hwang CH, Hwang BK (2008) Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227:409–425
Kamisugi Y, Cuming AC, Cove DJ (2005) Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Res 33(19):e173–e173
Kimber MS, Pai EF (2000) The active site architecture of Pisum sativum β-carbonic anhydrase is a mirror image of that of α-carbonic anhydrases. EMBO J 19(7):1407–1418
Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC (1996) A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J 15(10):2323–2330
Kravchik M, Bernstein N (2013) Effects of salinity on the transcriptome of growing maize leaf cells point at cell-age specificity in the involvement of the antioxidative response in cell growth restriction. BMC Genomics 14(1):24
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007) Clustal W and Clustal X version 20. Bioinformatics 23(21):2947–2948
Le BA, Scholz S, Kost B (2013) Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE 8(8):e70998
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46(D1):D493–D496
Mangani S, Håkansson K (1992) Crystallographic studies of the binding of protonated and unprotonated inhibitors to carbonic anhydrase using hydrogen sulphide and nitrate anions. Eur J Biochem 210(3):867–871
Moroney JV, Bartlett SG, Samuelsson G (2001) Carbonic anhydrases in plants and algae. Plant Cell Environ 24(2):141–153
Ortiz-Ramírez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L et al (2016) A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant 9(2):205–220
Parisi G, Perales M, Fornasari M, Colaneri A, Schain N, Casati D et al (2004) Gamma carbonic anhydrases in plant mitochondria. Plant Mol Biol 55(2):193–207
Price G, Howitt S, Harrison K, Badger M (1993) Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol 175:2871–2879
Prigge MJ, Bezanilla M (2010) Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137(21):3535–3543
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859):64–69
Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99:11640–11645
Studer AJ, Gandin A, Kolbe AR, Wang L, Cousins AB, Brutnell TP (2014) A limited role for carbonic anhydrase in C4 photosynthesis as revealed by a ca1ca2 double mutant in maize. Plant Physiol 165(2):608–617
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Wang C, Hu H, Qin X, Zeise B, Xu D, Rappel WJ et al (2016) Reconstitution of CO2 regulation of SLAC1 anion channel and function of CO2-permeable PIP2;1 aquaporin as CARBONIC ANHYDRASE4 interactor. Plant Cell 28(2):568–582
Wang M, Zhang Q, Liu FC, Xie WF, Wang GD, Wang J et al (2014) Family-wide expression characterization of Arabidopsis beta-carbonic anhydrase genes using qRT-PCR and promoter:GUS fusions. Biochimie 97:219–227
Wilkins MR, Gasteiger E, Bairoch A et al (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112(112):531–552
Yu S, Zhang X, Guan Q, Takano T, Liu S (2007) Expression of a carbonic anhydrase gene is induced by environmental stresses in rice (Oryza sativa L.). Biotechnol Lett 29(1):89–94
Zabaleta E, Martin MV, Braun HP (2012) A basal carbon concentrating mechanism in plants? Plant Sci 187:97–104
Zolfaghari Emameh R, Barker HR, Syrjänen L, Urbański L, Supuran CT, Parkkila S (2016) Identification and inhibition of carbonic anhydrases from nematodes. J Enzym Inhib Med Ch 31(sup4):176–184
Acknowledgements
We thank Prof. Mitsuyasu Hasebe for providing the moss spores and pTN182 plasmid. This work was supported by the CAS Pioneer Hundred Talents Program, the National Natural Science Foundation of China (31971410), the postdoctoral Fund of Yunnan Province (1ZY81C381261, Y732681261) and the open funds of the National Key Laboratory of Crop Genetic Improvement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare there have no conflict of interests.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Z., Wang, W., Dong, X. et al. Functional redundancy and divergence of β-carbonic anhydrases in Physcomitrella patens. Planta 252, 20 (2020). https://doi.org/10.1007/s00425-020-03429-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03429-8