Abstract
Main conclusion
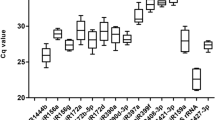
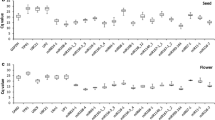
MicroRNAs miR390-5p, miR7694-3p miR1868 and miR1849 were found to be suitable miRNA reference genes for rice, under either infection with the root-knot nematode Meloidogyne graminicola or treatment with BABA.
Abstract
RT-qPCR is a widely used method to investigate the expression levels of genes under certain conditions. A key step, however, to have reliable results is the normalization of expression. For every experimental condition, suitable reference genes must be chosen. These reference genes must not be affected by differences in experimental conditions. MicroRNAs are regulatory RNA molecules, able to direct the expression levels of protein coding genes. In plants, their attributed functions range from roles in development to immunity. In this work, microRNAs (miRNAs) are evaluated for their suitability as reference genes in rice after infection with root-knot nematode Meloidogyne graminicola or after priming with beta-amino butyric acid. The evaluation was based on their amplification efficiency and their stability estimates according to geNorm, NormFinder and BestKeeper. All tested miRNAs, excluding one, were considered acceptable for normalization. Furthermore, miRNAs were validated using miRNA sequencing data. The set of microRNAs miR390-5p and miR7694-3p was found to be the most stable combination under the tested conditions. Another miRNA set consisting of miR7694-3p, miR1868 and miR1849 also shows potential to be used for miRNA expression normalization under experimental conditions beyond the scope of this study. This work is the first report on reference miRNAs in rice for the purpose of plant defence studies.



Similar content being viewed by others
Abbreviations
- BABA:
-
Beta-amino butyric acid
- CPM:
-
Counts per million mapped reads
- CV:
-
Coefficient of variation
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64VN-r:5245–5250. https://doi.org/10.1158/0008-5472.can-04-0496
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bridge J, Plowright RA, Peng D (2005) Nematode parasites of rice. In: Sikora R, Coyne D, Hallmann J, Timper P (eds) Plant parasitic nematodes in subtropical and tropical agriculture. CABI, Wallingford, pp 87–130
Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Buswell W, Schwarzenbacher RE, Luna E et al (2018) Chemical priming of immunity without costs to plant growth. New Phytol 218:1205–1216. https://doi.org/10.1111/nph.15062
Chugh P, Dittmer DP (2012) Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA 3:601–616. https://doi.org/10.1002/wrna.1120
Ding Y, Ye Y, Jiang Z et al (2016) MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci 7:235. https://doi.org/10.3389/fpls.2016.00235
Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA (2017) Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00378
Dobin A (2013) STAR for miRNA. https://groups.google.com/forum/#!topic/rna-star/RBWvAGFooMU. Accessed 14 May 2019
FAO (2017) Food and Agriculture Organization of the United Nations. http://www.fao.org/home/en. Accessed 28 May 2019
Fausto AKS, da Silva TF, Romanel E, Vaslin MFS (2017) MicroRNAs as reference genes for quantitative PCR in cotton. Public Libr Sci one 12:1–21. https://doi.org/10.1371/journal.pone.0174722
Gene Quantification Platform (2019) Real Time PCR amended, A useful new approach? Statistical problems? https://www.gene-quantification.de/avery-rel-pcr-errors.pdf. Accessed 12 Jun 2019
Gheysen G, Mitchum MG (2011) How nematodes manipulate plant development pathways for infection. Curr Opin Plant Biol 14:415–421. https://doi.org/10.1016/J.PBI.2011.03.012
Gutierrez L, Mauriat M, Gunin S et al (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618. https://doi.org/10.1111/j.1467-7652.2008.00346.x
Hellemans J, Mortier G, De Paepe A et al (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. https://doi.org/10.1186/gb-2007-8-2-r19
Ji H, Kyndt T, He W et al (2015) β-Aminobutyric acid-induced resistance against root-knot nematodes in rice is based on increased basal defense. Mol Plant-Microbe Interact 28:519–533. https://doi.org/10.1094/MPMI-09-14-0260-R
Jisha KC, Puthur JT (2016) Seed priming with BABA (β-amino butyric acid): a cost-effective method of abiotic stress tolerance in Vigna radiata (L.) Wilczek. Protoplasma 253:277–289. https://doi.org/10.1007/s00709-015-0804-7
Kozera B, Rapacz M (2013) Reference genes in real-time PCR. J Appl Genet 54:391–406. https://doi.org/10.1007/s13353-013-0173-x
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. https://doi.org/10.1093/nar/gky1141
Lawrence M, Huber W, Pagès H et al (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. https://doi.org/10.1371/journal.pcbi.1003118
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Lu Y, Feng Z, Liu X et al (2018) MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol 18:261. https://doi.org/10.1186/s12870-018-1488-x
Mestdagh P, Van Vlierberghe P, De Weer A et al (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. https://doi.org/10.1186/gb-2009-10-6-r64
Millar AA, Waterhouse PM (2005) Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5:129–135. https://doi.org/10.1007/s10142-005-0145-2
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. https://doi.org/10.1023/B:BILE.0000019559.84305.47
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369. https://doi.org/10.1038/nrg3198
Reversat G, Boyer J, Sannier C, Pando-Bahuon A (1999) Use of a mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1:209–212. https://doi.org/10.1163/156854199508027
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. https://doi.org/10.1186/gb-2010-11-3-r25
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81. https://doi.org/10.1016/S0165-022X(00)00129-9
Selvey S, Thompson EW, Matthaei K et al (2001) β-Actin—an unsuitable internal control for RT-PCR. Mol Cell Probes 15:307–311. https://doi.org/10.1006/MCPR.2001.0376
Slaughter A, Daniel X, Flors V et al (2012) Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 158:835–843. https://doi.org/10.1104/pp.111.191593
Vandesompele J, De Preter K, Pattyn I et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:34. https://doi.org/10.1186/gb-2002-3-7-research0034
Varkonyi-Gasic E, Wu R, Wood M et al (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:1–12. https://doi.org/10.1186/1746-4811-3-12
Wang ST, Sun XL, Hoshino Y et al (2014) MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS One 9:1–12. https://doi.org/10.1371/journal.pone.0091357
Yang C, Li D, Mao D et al (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36:2207–2218. https://doi.org/10.1111/pce.12130
Yang J, Zhang F, Li J et al (2016) Integrative analysis of the microRNAome and transcriptome illuminates the response of susceptible rice plants to rice stripe virus. PLoS One 11:e0146946. https://doi.org/10.1371/journal.pone.0146946
Zaleski C, Jha S, Gingeras TR et al (2012) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Zhang Y-C, Yu Y, Wang C-Y et al (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31:848–852. https://doi.org/10.1038/nbt.2646
Acknowledgements
This work was funded by Fonds Wetenschappelijk Onderzoek-Vlaanderen (Project: G007417N).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Verstraeten, B., De Smet, L., Kyndt, T. et al. Selection of miRNA reference genes for plant defence studies in rice (Oryza sativa). Planta 250, 2101–2110 (2019). https://doi.org/10.1007/s00425-019-03289-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03289-x