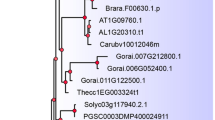
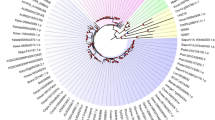
Abstract
Main conclusion
The work offers a comprehensive evaluation on the phylogenetics and conservation of splicing patterns of the plant SPF30 splicing factor gene family.
In eukaryotes, one pre-mRNA can generate multiple mRNA transcripts by alternative splicing (AS), which expands transcriptome and proteome diversity. Splicing factor 30 (SPF30), also known as survival motor neuron domain containing protein 1 (SMNDC1), is a spliceosomal protein that plays an essential role in spliceosomal assembly. Although SPF30 genes have been well characterised in human and yeast, little is known about their homologues in plants. Here, we report the genome-wide identification and phylogenetic analysis of SPF30 genes in the plant kingdom. In total, 82 SPF30 genes were found in 64 plant species from algae to land plants. Alternative transcripts were found in many SPF30 genes and splicing profile analysis revealed that the second intron in SPF30 genome is frequently associated with AS events and contributed to the birth of novel exons in a few SPF30 members. In addition, different conserved sequences were observed at these putative splice sites among moss, monocots and dicots, respectively. Our findings will facilitate further functional characterization of plant SPF30 genes as putative splicing factors.







Similar content being viewed by others
References
Abascal F, Tress ML, Valencia A (2015) The evolutionary fate of alternatively spliced homologous exons after gene duplication. Genome Biol Evol 7:1392–1403
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Alekseyenko AV, Kim N, Lee CJ (2007) Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA 13:661–670
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Barta A, Marquez Y, Brown JW (2012) Challenges in plant alternative splicing. In: Stamm S, Smith C, Lührmann R (eds) Alternative pre-mRNA splicing: theory and protocols. Wiley, New York, pp 79–91
Bernard P, Drogat J, Dheur S, Genier S, Javerzat JP (2010) Splicing factor Spf30 assists exosome-mediated gene silencing in fission yeast. Mol Cell Biol 30:1145–1157
Blencowe BJ (2017) The relationship between alternative splicing and proteomic complexity. Trends Biochem Sci 42:407–408
Buhler D, Raker V, Luhrmann R, Fischer U (1999) Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet 8:2351–2357
Bush SJ, Chen L, Tovar-Corona JM, Urrutia AO (2017) Alternative splicing and the evolution of phenotypic novelty. Philos T R Soc B 372:20150474
Caceres JF, Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18:186–193
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST plus: architecture and applications. BMC Bioinformatics 10:421
Chamala S, Feng G, Chavarro C, Barbazuk WB (2015) Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front Bioeng Biotechnol 3:33
Corpet F (1988) Multiple sequence alignment with hierarchical-clustering. Nucleic Acids Res 16:10881–10890
Cote J, Richard S (2005) Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280:28476–28483
Darracq A, Adams KL (2013) Features of evolutionarily conserved alternative splicing events between Brassica and Arabidopsis. New Phytol 199:252–263
Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Johnson LS, Eddy SR, Portugaly E (2010) Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431
Kalyna M, Lopato S, Voronin V, Barta A (2006) Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res 34:4395–4405
Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–355
Kim E, Goren A, Ast G (2008) Alternative splicing: current perspectives. BioEssays 30:38–47
Kopelman NM, Lancet D, Yanai I (2005) Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 37:588–589
Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Bio 14:153–165
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Lewis BP, Green RE, Brenner SE (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. P Natl Acad Sci USA 100:189–192
Little JT, Jurica MS (2008) Splicing factor SPF30 bridges an interaction between the prespliceosome protein U2AF35 and tri-small nuclear ribonucleoprotein protein hPrp3. J Biol Chem 283:8145–8152
Liu K, Guo YH, Liu HP, Bian CB, Lam R, Liu YS, Mackenzie F, Rojas LA, Reinberg D, Bedford MT, Xu RM, Min JR (2012) Crystal Structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One 7:e30375
Lu TT, Lu GJ, Fan DL, Zhu CR, Li W, Zhao QA, Feng Q, Zhao Y, Guo YL, Li WJ, Huang XH, Han B (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20:1238–1249
Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22:1184–1195
Matera AG, Wang ZF (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Bio 15:108–121
Mei WB, Boatwright L, Feng GQ, Schnable JC, Barbazuk WB (2017) Evolutionarily conserved alternative splicing across monocots. Genetics 207:465–480
Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U (2001) SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J 20:2304–2314
Morariu VI, Srinivasan BV, Raykar VC, Duraiswami R, Davis LS (2008) Automatic online tuning for fast Gaussian summation. Advances in Neural Information Processing Systems, 21st conference. pp 1113–1120
Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet 20:46–50
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415
Rappsilber J, Ajuh P, Lamond AI, Mann M (2001) SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J Biol Chem 276:31142–31150
Reddy ASN (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58:267–294
Schlaen RG, Mancini E, Sanchez SE, Perez-Santangelo S, Rugnone ML, Simpson CG, Brown JWS, Zhang X, Chernomoretz A, Yanovsky MJ (2015) The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. P Natl Acad Sci USA 112:9382–9387
Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M (2001) SMN Tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol 8:27–31
Sorek R (2007) The birth of new exons: mechanisms and evolutionary consequences. RNA 13:1603–1608
Staiger D, Brown JWS (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25:3640–3656
Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting CP, Davies KE (1998) Characterization of a gene encoding survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum Mol Genet 7:2149–2156
Tian Y, Chen M-X, Yang J-F, Achala H, Gao B, Hao G-F, Yang G-F, Dian Z-Y, Hu Q-J, Zhang D (2018) Genome-wide identification and functional analysis of the splicing component SYF2/NTC31/p29 across different plant species. Planta 1-18
Tress ML, Abascal F, Valencia A (2017) Most alternative isoforms are not functionally important. Trends Biochem Sci 42:408–410
Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M (2011) Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 18:1414
Wang W, Zheng H, Yang S, Yu H, Li J, Jiang H, Su J, Yang L, Zhang J, McDermott J, Samudrala R, Wang J, Yang H, Yu J, Kristiansen K, Wong GK, Wang J (2005) Origin and evolution of new exons in rodents. Genome Res 15:1258–1264
Wang BB, O’Toole M, Brendel V, Young ND (2008a) Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol 8:17
Wang ET, Sandberg R, Luo SJ, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008b) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476
Will CL, Luhrmann R (2011) Spliceosome structure and function. Csh Perspect Biol 3:a003707
Xing Y, Lee C (2006) Alternative splicing and RNA selection pressure—evolutionary consequences for eukaryotic genomes. Nat Rev Genet 7:499–509
Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13:1402–1406
Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133:585–600
Zhu FY, Chen MX, Ye NH, Shi L, Ma KL, Yang JF, Cao YY, Zhang YJ, Yoshida T, Fernie AR, Fan GY, Wen B, Zhou R, Liu TY, Fan T, Gao B, Zhang D, Hao GF, Xiao S, Liu YG, Zhang JH (2017) Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J 91:518–533
Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong Province (2018A030313030) and Hong Kong Research Grant Council (AoE/M-05/12, AoE/M-403/16, GRF14160516, 14177617, 12100318).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Phylogenetic tree of plant SPF30 gene family constructed by Maximum likelihood methods.
Figure S2 Weblogo plots of conserved motifs in plant SPF30 family.
Table S1SPF30 genes identified from 64 plant species.
Table S2 Predicted subcellular localization of plant SPF30 proteins.
Table S3 Characteristics of plant SPF30 gene structures.
Rights and permissions
About this article
Cite this article
Zhang, D., Yang, JF., Gao, B. et al. Identification, evolution and alternative splicing profile analysis of the splicing factor 30 (SPF30) in plant species. Planta 249, 1997–2014 (2019). https://doi.org/10.1007/s00425-019-03146-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03146-x