Abstract
Main conclusion
The LysM receptor-like kinase K1 is involved in regulation of pea-rhizobial symbiosis development.
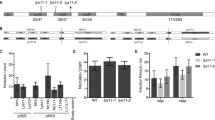
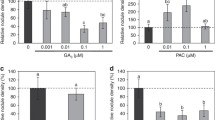
The ability of the crop legume Pisum sativum L. to perceive the Nod factor rhizobial signals may depend on several receptors that differ in ligand structure specificity. Identification of pea mutants defective in two types of LysM receptor-like kinases (LysM-RLKs), SYM10 and SYM37, featuring different phenotypic manifestations and impaired at various stages of symbiosis development, corresponds well to this assumption. There is evidence that one of the receptor proteins involved in symbiosis initiation, SYM10, has an inactive kinase domain. This implies the presence of an additional component in the receptor complex, together with SYM10, that remains unknown. Here, we describe a new LysM-RLK, K1, which may serve as an additional component of the receptor complex in pea. To verify the function of K1 in symbiosis, several P. sativum non-nodulating mutants in the k1 gene were identified using the TILLING approach. Phenotyping revealed the blocking of symbiosis development at an appropriately early stage, strongly suggesting the importance of LysM-RLK K1 for symbiosis initiation. Moreover, the analysis of pea mutants with weaker phenotypes provides evidence for the additional role of K1 in infection thread distribution in the cortex and rhizobia penetration. The interaction between K1 and SYM10 was detected using transient leaf expression in Nicotiana benthamiana and in the yeast two-hybrid system. Since the possibility of SYM10/SYM37 complex formation was also shown, we tested whether the SYM37 and K1 receptors are functionally interchangeable using a complementation test. The interaction between K1 and other receptors is discussed.









Similar content being viewed by others
Abbreviations
- ECD:
-
Extracellular domain
- GUS:
-
Beta-glucuronidase
- HR:
-
Hypersensitivity reaction
- IT:
-
Infection thread
- RLK:
-
Receptor-like kinase
- Y2H:
-
Yeast two-hybrid system
References
Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC et al (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6:1357–1374. https://doi.org/10.1105/tpc.6.10.1357
Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142:265–279. https://doi.org/10.1104/pp.106.084657
Ben Amor B, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D et al (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34:495–506. https://doi.org/10.1046/j.1365-313X.2003.01743.x
Bensmihen S, De Billy F, Gough C (2011) Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE 6:e26114. https://doi.org/10.1371/journal.pone.0026114
Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO et al (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131(3):1009–1017. https://doi.org/10.1104/pp.102.016071
Borisov AY, Vasilchikov AG, Voroshilova VA, Danilova TN, Zhernakov AI, Zhukov VA et al (2007) Regulatory genes of garden pea (Pisum sativum L.) controlling the development of nitrogen-fixing nodules and arbuscular mycorrhiza: a review of basic and applied aspects. Appl Biochem Microbiol 43:237–243. https://doi.org/10.1134/S0003683807030027
Brewin NJ, Wood EA, Larkins AP, Galfre G, Butcher GW (1986) Analysis of lipopolysaccharide from root nodule bacteroids of Rhizobium leguminosarum using monoclonal antibodies. J Gen Microbiol 132:1959–1968
Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N et al (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109(34):13859–13864. https://doi.org/10.1073/pnas.1205171109
Cao Y, Liang Y, Tanaka K, Nguyen C, Jedrzejczak R, Joachimiak A et al (2014) The kinase LYK5 is a major chitin receptor in and forms a chitin-induced complex with related kinase CERK1. Elife 3:e3766. https://doi.org/10.7554/eLife.03766
Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D et al (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128:1507–1518
Cullimore J, Dénarié J (2003) How legumes select their sweet talking symbionts. Science 302:575–578. https://doi.org/10.1126/science.1091269
Cullimore JV, Ranjeva R, Bono JJ (2001) Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci 6:24–30. https://doi.org/10.1016/S1360-1385(00)01810-0
Dalmais M, Schmidt J, Le Signor C, Moussy F, Burstin J, Savois V et al (2008) UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol 9:R43. https://doi.org/10.1186/gb-2008-9-2-r43
Dénarié J, Cullimore J (1993) Lipo-oligosaccharide nodulation factors: a minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 74:951–954. https://doi.org/10.1016/0092-8674(93)90717-5
Dénarié J, Debelle F, Prome JC (1996) Rhizobium lipo-chitooligosaccaride nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65:503–535. https://doi.org/10.1146/annurev.bi.65.070196.002443
Duc G, Messager A (1989) Mutagenesis of pea (Pisum sativum L.) and the isolation of mutants for nodulation and nitrogen fixation. Plant Sci 60:207–213. https://doi.org/10.1016/0168-9452(89)90168-4
Economou A, Davies AE, Johnston AWB, Downie JA (1994) The Rhizobium leguminosarum biovar viciae nodO gene can enable a nodE mutant of Rhizobium leguminosarum biovar trifolii to nodulate vetch. Microbiology 140:2341–2347
Engvild KC (1987) Nodulation and nitrogen fixation mutants of pea Pisum sativum. Theor Appl Genet 74:711–713. https://doi.org/10.1007/BF00247546
Fliegmann J, Canova S, Lachaud C, Uhlenbroich S, Gasciolli V, Pichereaux C et al (2013) Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem Biol 9:1900–1906. https://doi.org/10.1021/cb400369u
Geurts R, Heidstra R, Hadri A-E, Downie A, Franssen H, van Kammen A et al (1997) Sym2 of Pisum sativum is involved in Nod factor perception mechanism that controls the infection process in the epidermis. Plant Physiol 115:351–359. https://doi.org/10.1104/pp.115.2.351
Heidstra R, Bisseling T (1996) Nod factor-induced host responses and mechanisms of Nod factor perception. New Phytol 133:25–43. https://doi.org/10.1111/j.1469-8137.1996.tb04339.x
Indrasumunar A, Kereszt A, Searle I, Miyagi M, Li D, Nguyen CDT et al (2010) Inactivation of duplicated Nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol 51:201–214. https://doi.org/10.1093/pcp/pcp178
Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyam C, Dohmae N, Takio K et al (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103:11086–11091. https://doi.org/10.1073/pnas.0508882103
Kelly S, Radutoiu S, Stougaard J (2017) Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr Opin Plant Biol 39:152–158. https://doi.org/10.1016/j.pbi.2017.06.013
Kitaeva A, Demchenko K, Tikhonovich I, Timmers A, Tsyganov V (2016) Comparative analysis of the tubulin cytoskeleton organization in nodules of Medicago truncatula and Pisum sativum: bacterial release and bacteroid positioning correlate with characteristic microtubule rearrangements. New Phytol 210:168–183. https://doi.org/10.1111/nph.13792
Klaus-Heisen D, Nurisso A, Pietraszewska-Bogiel A, Mbengue M, Camut S, Timmers T et al (2011) Structure-function similarities between a plant receptor-like kinase and the human interleukin-1 receptor-associated kinase-4. J Biol Chem 286(13):11202–11210. https://doi.org/10.1074/jbc.M110.186171
Kneen B, Weeden N, LaRue T (1994) Non-nodulating mutants of Pisum sativum (L.) cv. Sparkle. J Heredity 85:129–133. https://doi.org/10.1093/oxfordjournals.jhered.a111410
Kouzai Y, Mochizuki S, Nakajima K, Desaki Y, Hayafune M, Miyazaki H et al (2014) Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol Plant Microbe Interact 27:975–982. https://doi.org/10.1094/MPMI-03-14-0068-R
Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C (2002) Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 99:11405–11410. https://doi.org/10.1073/pnas.172390699
Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome J-C et al (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781–784. https://doi.org/10.1038/344781a0
Lie TA (1984) Host genes in Pisum sativum L. conferring resistance to European Rhizobium leguminosarum strains. Plant Soil 82:415–425. https://doi.org/10.1007/BF02184279
Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302:630–633. https://doi.org/10.1126/science.1090074
Liu B, Li JF, Ao Y, Qu J, Li Z, Su J et al (2012a) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24:3406–3419. https://doi.org/10.1105/tpc.112.102475
Liu T, Liu Z, Song C, Hu Y, Han Z, She J et al (2012b) Chitin-induced dimerization activates a plant immune receptor. Science 336:1160–1163. https://doi.org/10.1126/science.1218867
Lohmann GV, Shimoda Y, Nielsen MW, Jørgensen FG, Grossmann C, Sandal N et al (2010) Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact 23(4):510–521. https://doi.org/10.1094/MPMI-23-4-0510
Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425:637–640. https://doi.org/10.1038/nature02045
Madsen L, Tirichine L, Jurkiewicz A, Sullivan J, Heckmann A, Bek A et al (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1:1–12. https://doi.org/10.1038/ncomms1009
Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A et al (2011) Autophosphorylation is essential for in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor mediated signalling in cooperation with Nod factor receptor 5. Plant J 65:404–417. https://doi.org/10.1111/j.1365-313X.2010.04431.x
Mergaert P, Van Montagu M, Holsters M (1997) Molecular mechanisms of Nod factor diversity. Mol Microbiol 25:811–817. https://doi.org/10.1111/j.1365-2958.1997.mmi526.x
Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K et al (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104:19613–19618. https://doi.org/10.1073/pnas.0705147104
Miyata K, Kozaki T, Kouzai Y, Ozawa K, Ishii K, Asamizu E et al (2014) The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol 55:1864–1872. https://doi.org/10.1093/pcp/pcu129
Moling S, Pietraszewska-Bogiel A, Postma M, Fedorova E, Hink MA, Limpens E et al (2014) Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26:4188–4199. https://doi.org/10.1105/tpc.114.129502
Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanashi K et al (2011) From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J 65:169–180. https://doi.org/10.1111/j.1365-313X.2010.04411.x
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812-3814. PMCID: PMC168916
Oldroyd GE, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago trancatula. Plant Cell 13:1835–1849. https://doi.org/10.1105/TPC.010193
Orosz L, Sváb Z, Kondorosi A, Sik T (1973) Genetic studies on rhizobiophage 16-3. I. Genes and functions on the chromosome. Mol Gen Genet 125:341–350. https://doi.org/10.1007/BF00276589
Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbial Mol Biol Rev 64:180–201
Pietraszewska-Bogiel A, Lefebvre B, Koini MA, Klaus-Heisen D, Takken FL, Geurts R et al (2013) Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS ONE 8:650–655. https://doi.org/10.1371/journal.pone.0065055
Postma JG, Jacobsen E, Feenstra WJ (1988) Three pea mutants with an altered nodulation studied by genetic analysis and grafting. J Plant Physiol 132:424–430. https://doi.org/10.1016/S0176-1617(88)80056-7
Prell J (2003) Physiologische und genetische Charakterisierung der g-Amino-butyrat (GABA) aminotransferase in Rhizobium leguminosarum bv. viciae VF39: Dissertation, Bibliothek der RWTH Aachen
Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585–592. https://doi.org/10.1038/nature02039
Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM et al (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26:3923–3935. https://doi.org/10.1038/sj.emboj.7601826
Riely BK, Ané JM, Penmetsa RV, Cook DR (2004) Genetic and genomic analysis in model legumes bring Nod factor signaling to center stage. Curr Opin Plant Biol 7:408–413. https://doi.org/10.1016/j.pbi.2004.04.005
Sagan OM, Huguet T, Duc G (1994) Phenotypic characterization and classification of nodulation mutants of pea (Pisum sativum L.). Plant Sci 100:59–70. https://doi.org/10.1016/0168-9452(94)90134-1
Schauser L, Rouss A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402:191–195. https://doi.org/10.1038/46058
Schneider A, Walker SA, Poyser S, Sagan M, Ellis TH, Downie JA (1999) Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol Gen Genet 262:1–11. https://doi.org/10.1007/s004380051053
Schneider A, Walker SA, Sagan M, Duc G, Ellis THN, Downie JA (2002) Mapping of the nodulation loci sym9 and sym10 of pea (Pisum sativum L.). Theor Appl Genet 104:1312–1316. https://doi.org/10.1007/s00122-002-0896-2
Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32:33–57. https://doi.org/10.1146/annurev.genet.32.1.33
Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y et al (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64:204–214. https://doi.org/10.1111/j.1365-313X.2010.04324.x
Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C et al (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145:183–191. https://doi.org/10.1104/pp.107.100495
Spaink HP, Sheeley DM, van Brussel AAN, Glushka J, York WS, Tak T et al (1991) A novel highly saturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354:125–130. https://doi.org/10.1038/354125a0
Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962. https://doi.org/10.1038/nature00841
Tsyganov V, Voroshilova V, Priefer U, Borisov A, Tikhonovich I (2002) Genetic dissection of the initiation of the infection process and nodule tissue development in the Rhizobium-pea (Pisum sativum L.) symbiosis. Ann Bot. 89:357–366. https://doi.org/10.1093/aob/mcf051
Tsyganova AV, Tsyganov VE, Findlay KC, Borisov AY, Tikhonovich IA, Brewin NJ (2009) Distribution of legume arabinogalactan protein-extensin (AGPE) glycoproteins in symbiotically defective pea mutants with abnormal infection threads. Cell Tissue Biol 3:93–102. https://doi.org/10.1134/S1990519X09010131
van Brussel AAN, Planque K, Quispel A (1977) The wall of Rhizobium leguminosarum in bacteroid and free-living forms. J Gen Microbiol 101:51–56. https://doi.org/10.1099/00221287-101-1-51
van Brussel AAN, Tak T, Wetselaar A, Pees E, Wijffelman CA (1982) Small leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci Lett 27:317–325. https://doi.org/10.1016/0304-4211(82)90134-1
van den Bosch KA, Bradley DJ, Knox JP, Perotto S, Butcher GW, Brewin NJ (1989) Common components of the infection thread matrix and intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J 8:335–342
van Spronsen PC, Bakhuizen R, van Brussel AA, Kijne JW (1994) Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. Eur J Cell Biol 4(1):88–94
Voroshilova VA, Demchenko KN, Brewin NJ, Borisov AY, Tikhonovich IA (2009) Initiation of a legume nodule with an indeterminate meristem involves proliferating host cells that harbour infection threads. New Phytol 181:913–923. https://doi.org/10.1111/j.1469-8137.2008.02723.x
Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D et al (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97:13407–13412. https://doi.org/10.1073/pnas.230439797
Walker SA, Downie JA (2000) Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal Nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol Plant Microbe Interact 13:754–762. https://doi.org/10.1094/MPMI.2000.13.7.754
Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K et al (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108:19824–19829. https://doi.org/10.1073/pnas.1112862108
Zhang X-C, Wu X, Findley S, Wan J, Libault M, Nguyen HT et al (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144:623–636. https://doi.org/10.1104/pp.107.097097
Zhukov V, Radutoiu S, Madsen LH, Rychagova T, Ovchinnikova E, Borisov A et al (2008) The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol Plant Microbe Interact 21:1600–1608. https://doi.org/10.1094/MPMI-21-12-1600
Acknowledgements
This work was funded for ARRIAM (A.N.K., N.V.M., G.A.A., I.A.T., and E.A.D.) by Russian Scientific Foundation (RSF project no. 16-16-10043) and for ITMO University (Yu.B.P) by Government of Russian Federation, Grant 074-U01. The INRA TILLING activities are supported by the Program Saclay Plant Sciences (SPS, ANR-10-LABX-40) and the European Research Council (ERC-SEXYPARTH). The research was performed using the equipment of the Core Center “Genomic technologies, proteomics and cellular biology” at ARRIAM. We are very grateful to Dr. Julie Cullimore (Laboratory of Plant–Microbe Interactions, INRA, France) for kindly providing the constructs for production of AtCERK1-ECD in yeast and for the pMON and pBIN vectors. Dr. Zhukov V. and Sulima A. for providing K1 and Sym37 sequences (cv. Cameor). Kitaeva A. for helping in microscopy analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
No conflicts of interest declared.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kirienko, A.N., Porozov, Y.B., Malkov, N.V. et al. Role of a receptor-like kinase K1 in pea Rhizobium symbiosis development. Planta 248, 1101–1120 (2018). https://doi.org/10.1007/s00425-018-2944-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-018-2944-4