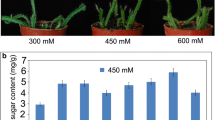
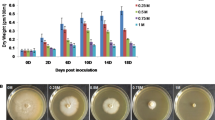
Abstract
Main Conclusion
Ribosome activation and sugar metabolic process mainly act on the regulation of salt tolerance in the bioenergy crop Helianthus tuberosus L. as dissected by integrated transcriptomic and proteomic analyses.
Helianthus tuberosus L. is an important halophyte plant that can survive in saline–alkali soil. It is vitally necessary to build an available genomic resource to investigate the molecular mechanisms underlying salt tolerance in H. tuberosus. De novo assembly and annotation of transcriptomes were built for H. tuberosus using a HiSeq 4000 platform. 293,823 transcripts were identified and annotated into 190,567 unigenes. In addition, iTRAQ-labeled quantitative proteomics was carried out to detect global protein profiling as a response to salt stress. Comparative omics analysis showed that 5432 genes and 43 proteins were differentially expressed in H. tuberosus under salt stress, which were enriched in the following processes: carbohydrate metabolism, ribosome activation and translation, oxidation–reduction and ion binding. The reprogramming of transcript and protein works suggested that the induced activity of ribosome and sugar signaling may endue H. tuberosus with salt tolerance. With high-quality sequencing and annotation, the obtained transcriptomics and proteomics provide a robust genomic resource for dissecting the regulatory molecular mechanism of H. tuberosus in response to salt stress.









Similar content being viewed by others
Abbreviations
- DEPC:
-
Diethy pyrocarbonate
- DEGs:
-
Differentially expressed genes
- FPKM:
-
Fragments per kilobase per transcript per million mapped reads
- MDHAR:
-
Monodehydroascorbate reductase
- LEA:
-
Late embryogenesis abundant
- RSEM:
-
RNA-seq by expectation maximization
References
Abogadallah GM (2010) Antioxidative defense under salt stress. Plant Signal Behav 5:369–374
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. https://doi.org/10.1186/gb-2010-11-10-r106
Asensi-Fabado MA, Ammon A, Sonnewald U, Munné-Bosch S, Voll LM (2015) Tocopherol deficiency reduces sucrose export from salt-stressed potato leaves independently of oxidative stress and symplastic obstruction by callose. J Exp Bot 66:957–971. https://doi.org/10.1093/jxb/eru453
Baack EJ, Whitney KD, Rieseberg LH (2005) Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol 167:623–630. https://doi.org/10.1111/j.1469-8137.2005.01433.x
Baiya S, Hua Y, Ekkhara W, Ketudat Cairns JR (2014) Expression and enzymatic properties of rice (Oryza sativa L.) monolignol β-glucosidases. Plant Sci 227:101–109. https://doi.org/10.1016/j.plantsci.2014.07.009
Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127:398–415. https://doi.org/10.1104/pp.127.2.398
Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH (2008) Multiple paleopolyploidizations during the evolution of the Asteraceae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol 25:2445–2455. https://doi.org/10.1093/molbev/msn187
Barta J, Pátkai GY (2007) Chemical composition and storability of Jerusalem artichoke tubers. Acta Aliment Hung 36:257–267. https://doi.org/10.1556/AAlim.36.2007.2.13
Bock DG, Kane NC, Ebert DP, Rieseberg LH (2014) Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: neither from Jerusalem nor an artichoke. New Phytol 201:1021–1030. https://doi.org/10.1111/nph.12560
Boriboonkaset T, Theerawitaya C, Yamada N, Pichakum A, Supaibulwatana K, Cha-Um S, Takabe T, Kirdmanee C (2013) Regulation of some carbohydrate metabolism-related genes, starch and soluble sugar contents, photosynthetic activities and yield attributes of two contrasting rice genotypes subjected to salt stress. Protoplasma 250:1157–1167. https://doi.org/10.1007/s00709-013-0496-9
Chen Q, Zhang MD, Shen SH (2011) Effect of salt on malondialdehyde and antioxidant enzymes in seedling roots of Jerusalem artichoke (Helianthus tuberosus L.). Acta Physiol Plant 33:273–278. https://doi.org/10.1007/s11738-010-0543-5
de Sá PH, Veras AA, Carneiro AR, Pinheiro KC, Pinto AC, Soares SC, Schneider MP, Azevedo V, Silva A, Ramos RT (2015) The impact of quality filter for RNA-Seq. Gene 563:165–171. https://doi.org/10.1016/j.gene.2015.03.033
Draper HH, Hadley M (1990) Malondialdehyde determination as index of lipid peroxidation. Meth Enzymol 186:421–431. https://doi.org/10.1016/0076-6879(90)86135-i
Edelman J, Jefford TG (1968) The mechanism of fructosan metabolism in higher plants as exemplified in Helianthus tuberosus. New Phytol 67:517–531. https://doi.org/10.1111/j.1469-8137.1968.tb05480.x
Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228:191–201. https://doi.org/10.1007/s00425-008-0729-x
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. https://doi.org/10.1093/nar/gkn176
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
He W, Zhuang HH, Fu YP, Guo LW, Guo B, Guo LZ, Zhang XH, Wei YH (2015) De novo transcriptome assembly of a Chinese locoweed (Oxytropis ochrocephala) species provides insights into genes associated with drought, salinity, and cold tolerance. Front Plant Sci 6:1086. https://doi.org/10.3389/fpls.2015.01086
Henry C, Bledsoe SW, Griffiths CA, Kollman A, Paul MJ, Sakr S, Lagrimini LM (2015) Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol 169:1072–1089. https://doi.org/10.1104/pp.15.00729
Higashiyama T (2002) Novel functions and applications of trehalose. Pure Appl Chem 74:1263–1269. https://doi.org/10.1351/pac200274071263
Huang ZR, Long XH, Wang L, Kang J, Zhang ZH, Zed R, Liu ZP (2012) Growth, photosynthesis and H+-ATPase activity in two Jerusalem artichoke varieties under NaCl-induced stress. Process Biochem 47:591–596. https://doi.org/10.1016/j.procbio.2011.12.016
Huang ZR, Zhao L, Chen DD, Liang MX, Liu ZP, Shao HB, Long XH (2013) Salt stress encourages proline accumulation by regulating proline biosynthesis and degradation in Jerusalem artichoke plantlets. PLoS One 8:e62085. https://doi.org/10.1371/journal.pone.0062085
Ibrahim MH, Jaafar HZ (2012) Primary, secondary metabolites, H2O2, malondialdehyde and photosynthetic responses of Orthosiphon stamineus Benth. to different irradiance levels. Molecules 17:1159–1176. https://doi.org/10.3390/molecules17021159
Jin HX, Dong DK, Yang QH, Zhu DH (2016) Salt-responsive transcriptome profiling of Suaeda glauca via RNA sequencing. PLoS One 11:e0150504. https://doi.org/10.1371/journal.pone.0150504
Jung WY, Lee SS, Kim CW, Kim HS, Min SR, Moon JS, Kwon SY, Jeon JH, Cho HS (2014) RNA-Seq analysis and de novo transcriptome assembly of Jerusalem artichoke (Helianthus tuberosus Linne). PLoS One 9:e111982. https://doi.org/10.1371/journal.pone.0111982
Kays SJ, Nottingham SF (2008) Biology and chemistry of the Jerusalem Artichoke: Helianthus tuberosus L. CRC Press, Boca Raton
Kocsy G, Tari I, Vanková R, Zechmann B, Gulyás Z, Poór P, Galiba G (2013) Redox control of plant growth and development. Plant Sci 211:77–91. https://doi.org/10.1016/j.plantsci.2013.07.004
Krasensky J, Broyart C, Rabanal FA, Jonak C (2014) The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid Redox Signal 21:1289–1304. https://doi.org/10.1089/ars.2013.5693
Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR (2012) A high-throughput method for Illumina RNA-Seq library preparation. Front Plant Sci 3:202. https://doi.org/10.3389/fpls.2012.00202
Lee BH, Eskandari R, Jones K, Reddy KR, Quezada-Calvillo R, Nichols BL, Rose DR, Hamaker BR, Pinto BM (2012) Modulation of starch digestion for slow glucose release through “toggling” of activities of mucosal α-glucosidases. J Biol Chem 287:31929–31938. https://doi.org/10.1074/jbc.M112.351858
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Li RX, Sun RB, Hicks GR, Raikhel NV (2015) Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc Natl Acad Sci USA 112:E89–E98. https://doi.org/10.1073/pnas.1422656112
Liu ZB, Li Y, Cao HW, Ren DT (2015) Comparative phospho-proteomics analysis of salt-responsive phosphoproteins regulated by the MKK9-MPK6 cascade in Arabidopsis. Plant Sci 241:138–150. https://doi.org/10.1016/j.plantsci.2015.10.005
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Long XH, Huang ZR, Huang YL, Kang J, Zhang ZH, Liu ZP (2010a) Response of two Jerusalem artichoke (Helianthus tuberosus) cultivars differing in tolerance to salt treatment. Pedosphere 20:515–524. https://doi.org/10.1016/S1002-0160(10)60041-0
Long XH, Huang ZR, Zhang ZH, Li Q, Zed R, Liu ZP (2010b) Seawater stress differentially affects germination, growth, photosynthesis, and ion concentration in genotypes of Jerusalem artichoke (Helianthus tuberosus L.). J Plant Growth Regul 29:223–231. https://doi.org/10.1007/s00344-009-9125-4
Ma XY, Zhang LH, Shao HB, Xu G, Zhang F, Ni FT, Brestic M (2011) Jerusalem artichoke (Helianthus tuberosus), a medicinal salt-resistant plant has high adaptability and multiple-use values. J Med Plant Res 5:1272–1279
Mao X, Cai T, Olyarchuk JG, Wei LP (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Miao ZY, Xu W, Li DF, Hu XN, Liu JX, Zhang RX, Tong ZY, Dong JL, Su Z, Zhang LW, Sun M, Li WJ, Du ZL, Hu SN, Wang T (2015) De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress-responsive pathway. BMC Genom 16:818. https://doi.org/10.1186/s12864-015-2019-x
Mohsenzadeh F, Chehregani A, Yousefi N (2011) Effect of the heavy metals on developmental stages of ovule, pollen, and root proteins in Reseda lutea L. (Resedaceae). Biol Trace Elem Res 143:1777–1788. https://doi.org/10.1007/s12011-011-9009-x
Nieves-Cordones M, Al Shiblawi FR, Sentenac H (2016) Roles and transport of sodium and potassium in plants. Met Ions Life Sci 16:291–324. https://doi.org/10.1007/978-3-319-21756-7_9
Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 154:373–390. https://doi.org/10.1104/pp.110.158964
Olvera-Carrillo Y, Reyes JL, Covarrubias AA (2011) Late embryogenesis abundant proteins: versatile players in the plant adaptation to water limiting environments. Plant Signal Behav 6:586–589. https://doi.org/10.4161/psb.6.4.15042
Pang QY, Chen SX, Dai SJ, Chen YZ, Wang Y, Yan XF (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9:2584–2599. https://doi.org/10.1021/pr100034f
Park HJ, Kim WY, Yun DJ (2016) A new insight of salt stress signaling in plant. Mol Cells 39:447–459. https://doi.org/10.14348/molcells.2016.0083
Peroza EA, dos Santos Cabral A, Wan X, Freisinger E (2013) Metal ion release from metallothioneins: proteolysis as an alternative to oxidation. Metallomics 5:1204–1214. https://doi.org/10.1039/c3mt00079f
Pospíšil P (2014) The role of metals in production and scavenging of reactive oxygen species in photosystem II. Plant Cell Physiol 55:1224–1232. https://doi.org/10.1093/pcp/pcu053
Potters G, Horemans N, Jansen MA (2010) The cellular redox state in plant stress biology—a charging concept. Plant Physiol Biochem 48:292–300. https://doi.org/10.1016/j.plaphy.2009.12.007
Rampitsch C, Bykova NV (2012) The beginnings of crop phosphoproteomics: exploring early warning systems of stress. Front Plant Sci 3:144. https://doi.org/10.3389/fpls.2012.00144
Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, Sinha NR (2014) De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol 166:1186–1199. https://doi.org/10.1104/pp.113.234864
Richter JA, Erban A, Kopka J, Zörb C (2015) Metabolic contribution to salt stress in two maize hybrids with contrasting resistance. Plant Sci 233:107–115. https://doi.org/10.1016/j.plantsci.2015.01.006
Rogers C, Thompson T, Seiler GJ (1982) Sunflower species of the United States. National Sunflower Association, Bismark
Saengthongpinit W, Sajjaanantakul T (2005) Influence of harvest time and storage temperature on characteristics of inulin from Jerusalem artichoke (Helianthus tuberosus L.) tubers. Postharvest Biol Technol 37:93–100. https://doi.org/10.1016/j.postharvbio.2005.03.004
Shabala S, Wu H, Bose J (2015) Salt stress sensing and early signalling events in plant roots: current knowledge and hypothesis. Plant Sci 241:109–119. https://doi.org/10.1016/j.plantsci.2015.10.003
Skorupa M, Gołębiewski M, Domagalski K, Kurnik K, Abu Nahia K, Złoch M, Tretyn A, Tyburski J (2016) Transcriptomic profiling of the salt stress response in excised leaves of the halophyte Beta vulgaris ssp. maritime. Plant Sci 243:56–70. https://doi.org/10.1016/j.plantsci.2015.11.007
Spoel SH, van Ooijen G (2014) Circadian redox signaling in plant immunity and abiotic stress. Antioxid Redox Signal 20:3024–3039. https://doi.org/10.1089/ars.2013.5530
Sultana S, Khew CY, Morshed MM, Namasivayam P, Napis S, Ho CL (2012) Overexpression of monodehydroascorbate reductase from a mangrove plant (AeMDHAR) confers salt tolerance on rice. J Plant Physiol 169:311–318. https://doi.org/10.1016/j.jplph.2011.09.004
Sun Y, Kong X, Li C, Liu Y, Ding Z (2015) Potassium retention under salt stress is associated with natural variation in salinity tolerance among Arabidopsis accessions. PLoS One 10:e0124032. https://doi.org/10.1371/journal.pone.0124032
Thompson JE, Fry SC (2001) Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J 26:23–34. https://doi.org/10.1046/j.1365-313x.2001.01005.x
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Valluru R, Van Den Ende W (2008) Plant fructans in stress environments: emerging concepts and future prospects. J Exp Bot 59:2905–2916. https://doi.org/10.1093/jxb/ern164
Wang XC, Chang LL, Wang BC, Wang D, Li PH, Wang LM, Yi XP, Huang QX, Peng M, Guo AP (2013) Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol Cell Proteom 12:2174–2195. https://doi.org/10.1074/mcp.M112.022475
Weis BL, Kovacevic J, Missbach S, Schleiff E (2015) Plant-specific features of ribosome biogenesis. Trends Plant Sci 20:729–740. https://doi.org/10.1016/j.tplants.2015.07.003
Wright L, Wrench P, Hinde RW, Brady CJ (1977) Proline accumulation in tubers of Jerusalem artichoke. Aust J Plant Physiol 4:51–60. https://doi.org/10.1071/PP9770051
Yamaguchi T, Blumwald E (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10:615–620. https://doi.org/10.1016/j.tplants.2005.10.002
Yates SA, Swain MT, Hegarty MJ, Chernukin I, Lowe M, Allison GG, Ruttink T, Abberton MT, Jenkins G, Skøt L (2014) De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom 15:453. https://doi.org/10.1186/1471-2164-15-453
You Q, Yi X, Zhang K, Yao DX, Zhang XY, Wang QH, Zhao XH, Ling Y, Xu WY, Li FG, Su Z (2016) Co-expression network analyses identify functional modules associated with development and stress response in Gossypium arboreum. Sci Rep 6:38436. https://doi.org/10.1038/srep38436
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yruela I (2013) Transition metals in plant photosynthesis. Metallomics 5:1090–1109. https://doi.org/10.1039/c3mt00086a
Yu B, Li JN, Koh J, Dufresne C, Yang N, Qi SS, Zhang YX, Ma CQ, Duong BV, Chen SX, Li HY (2016) Quantitative proteomics and phosphoproteomics of sugar beet monosomic addition line M14 in response to salt stress. J Proteom 143:286–297. https://doi.org/10.1016/j.jprot.2016.04.011
Zhang T, Zhu MM, Zhu N, Strul JM, Dufresne CP, Schneider JD, Harmon AC, Chen SX (2016) Identification of thioredoxin targets in guard cell enriched epidermal peels using cysTMT proteomics. J Proteomics 133:48–53. https://doi.org/10.1016/j.jprot.2015.12.008
Zhao GM, Liu ZP, Chen MD, Kou WF (2006) Effect of saline aquaculture effluent on salt-tolerant Jerusalem artichoke (Helianthus tuberosus L.) in a semi-arid coastal area of China. Pedosphere 16:762–769. https://doi.org/10.1016/S1002-0160(06)60112-4
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71. https://doi.org/10.1016/S1360-1385(00)01838-0
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31470467) and the Fundamental Research Funds for the Central Universities (2572016DA05 and 2572016AA15).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2017_2818_MOESM1_ESM.pptx
Supplementary Fig. S1 Assembled transcripts of H. tuberosus search against Blast Nr. Plant species classification of the unigenes based on the best species similarity, distribution of e-value showed in blast analysis of de novo assembled transcriptomes, sequence similarity distribution of unigenes with the top Nr hits (PPTX 685 kb)
425_2017_2818_MOESM2_ESM.pptx
Supplementary Fig. S2 Annotation of all the unigenes from H. tuberosus transcriptome. Number of genes annotated by five representatively databases (Blast Nr, Swiss Prot, GO, Pfam, KO) was showed in Venn diagram (PPTX 105 kb)
425_2017_2818_MOESM4_ESM.pptx
Supplementary Fig. S4 Volcano plot of differentially expressed proteins in H. tuberosus root under salt stress. The differentially expressed proteins were set with 0.85≥fold change≥1.2 and P-value≤0.05 calculated by Student’s t-test (PPTX 73 kb)
425_2017_2818_MOESM10_ESM.xlsx
Supplementary Table S6 Fragments per kilobase per transcript per million mapped reads (FPKM) interval in three independent biological replicates of control (JA-RC/JA-TC) and salt treated samples (JA-RT/JA-TT) of H. tuberosus (XLSX 11 kb)
425_2017_2818_MOESM12_ESM.xlsx
Supplementary Table S8 Expression of the down-regulated transcripts in H. tuberosus root under salt stress (XLSX 243 kb)
Rights and permissions
About this article
Cite this article
Zhang, A., Han, D., Wang, Y. et al. Transcriptomic and proteomic feature of salt stress-regulated network in Jerusalem artichoke (Helianthus tuberosus L.) root based on de novo assembly sequencing analysis. Planta 247, 715–732 (2018). https://doi.org/10.1007/s00425-017-2818-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2818-1