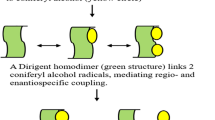
Abstract
Main conclusion
The plant LIMs comprise two sub-families with one (DA1/DAR) and two (2LIM) LIM domains. This review comprehensively discussed the structure and potential role of this protein family in diverse area of plant biology.
The description of first eukaryote lineage-specific plant LIM domain (LIN11, ISL1, and MEC3) proteins was observed in Helianthus long back. The successive study of LIM proteins in diverse plants has shown its vital relation to development, metabolism and defence. This nascent gene family has been worked out for their role in actin dynamics, organ size determination and transcription regulation. On grounds of protein architecture, two sub-families have been delineated as DA1/DAR (one LIM domain) and 2LIMs (two LIM domains). The genomic and expression study guides to the identification of diverse sub-categories. The significance of 2LIMs in regulation of actin dynamics leading to pollen growth and development has prospects to understand the plant reproductive behaviour. Interestingly, new facet of these LIMs as a transcriptional regulator in biological pathway/biosynthesis was also reported. Recently, the cumulative contribution of these features was also recognized for obtaining good quality fibre, thus giving translational outlook to this family. The DA1/DAR proteins are orchestrated with additional domains and provide a key role in regulation of organ size and tolerance to biotic and abiotic stress. This review will focus the journey of plant LIMs till date and will cover details of its structure, type, classification and functional relevance. This will provide insight to identify the potential of this gene family in the improvement of desired crop features.


Similar content being viewed by others
References
Arnaud D, Déjardin A, Leplé JC, Lesage-Descauses MC, Pilate G (2007) Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res 14:103–116
Arnaud D, Déjardin A, Leplé JC, Lesage-Descauses MC, Boizot N, Villar M, Bénédetti H, Pilate G (2012) Expression analysis of LIM gene family in poplar, toward an updated phylogenetic classification. BMC Res Notes 5:102
Baltz R, Evrard JL (1996) The pollen-specific LIM protein PLIM-1 from sunflower binds nucleic acids in vitro. Sex Plant Repro 9:264–268
Baltz R, Evrard JL, Domon C, Steinmetz A (1992) A LIM motif is present in a pollen-specific protein. Plant Cell 4:1465–1466
Baltz R, Schmit AC, Kohnen M, Hentges F, Steinmetz A (1999) Differential localization of the LIM domain protein PLIM-1 in microspores and mature pollen grains from sunflower. Sex Plant Reprod 12:60–65
Bi D, Johnson KC, Zhu Z, Huang Y, Chen F, Zhang Y, Li X (2011) Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front Plant Sci 2:71
Brière C, Bordel AC, Barthou H, Jauneau A, Steinmetz A, Alibert G, Petitprez M (2003) Is the LIM-domain protein HaWLIM1 associated with cortical microtubules in sunflower protoplasts? Plant Cell Physiol 44:1055–1063
Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26:665–677
Eliasson A, Gass N, Munde C, Baltz R, Krauter R, Evrard JL, Steinmetz A (2000) Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol Genet Genom 264:257–267
Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9:e0152
Freyd G, Kim SK, Horvitz HR (1990) Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344:876–879
Han LB, Li YB, Wang HY, Wu XM, Li CL, Luo M, Wu SJ, Kong ZS, Pei Y, Jiao GL, Xia GX (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25:4421–4438
Han L, Li Y, Sun Y, Wang H, Kong Z, Xia G (2016) The two domains of cotton WLIM1a protein are functionally divergent. Sci Chin Life Sci 59:206–212
Hobert O, Westphal H (2000) Functions of LIM-homeobox genes. Trends Genet 16:75–83
Hoffmann C, Moes D, Dieterle M, Neumann K, Moreau F, Furtado AT, Dumas D, Steinmetz A, Thomas C (2014) Live cell imaging reveals actin-cytoskeleton-induced self-association of the actin-bundling protein WLIM1. J Cell Sci 127:583–598
Kadrmas JL, Beckerle MC (2004) The LIM domain: from the cytoskeleton to the nucleus. Nature Rev Mol Cell Biol 5:920–931
Kaothien P, Kawaoka A, Ebinuma H, Yoshida K, Shinmyo A (2002) Ntlim1, a PAL-box binding factor, controls promoter activity of the horseradish wound-inducible peroxidase gene. Plant Mol Biol 49:591–599
Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T (1990) Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344:879–882
Kawaoka A, Ebinuma H (2001) Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry 57:1149–1157
Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22:289–301
Kawaoka A, Nanto K, Ishii K, Ebinuma H (2006) Reduction of lignin content by suppression of expression of the LIM domain transcription factor in Eucalyptus camaldulensis. Sil Genet 55:269–277
Khatun K, Robin AHK, Park JI, Ahmed NU, Kim CK, Lim KB, Kim MB, Lee DJ, Nou IS, Chung MY (2016) Genome-wide identification, characterization and expression profiling of LIM family genes in Solanum lycopersicum L. Plant Physiol Biochem 108:177–190
Khurana P, Henty JL, Huang S, Staiger AM, Blanchoin L, Staiger CJ (2010) Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22:2727–2748
Li N, Li Y (2015a) Maternal control of seed size in plants. J Exp Bot 66:1087–1097
Li N, Li Y (2015b) Ubiquitin-mediated control of seed size in plants. Front Plant Sci 5:332
Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22:1331–1336
Li Y, Jiang J, Li L, Wang XL, Wang NN, Li DD, Li XB (2013) A cotton LIM domain-containing protein (GhWLIM5) is involved in bundling actin filaments. Plant Physiol Biochem 66:34–40
Li Y, Yue X, Que Y, Yan X, Ma Z, Talbot NJ, Wang Z (2014) Characterisation of four LIM protein-encoding genes involved in infection-related development and pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS Osne 9:e88246
Li L, Li Y, Wang NN, Lu R, Li XB (2015) Cotton LIM domain-containing protein GhPLIM1 is specifically expressed in anthers and participates in modulating F-actin. Plant Biol 17:528–534
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol plant 8:689–708
Matthews JM, Lester K, Joseph S, Curtis DJ (2013) LIM-domain-only proteins in cancer. Nat Rev Can 13:111–122
Moes D, Gatti S, Hoffmann C, Dieterle M, Moreau F, Neumann K, Schumacher M, Diederich M, Grill E, Shen WH, Steinmetz A, Thomas C (2013) A LIM domain protein from tobacco involved in actin-bundling and histone gene transcription. Mol Plant 6:483–502
Moes D, Hoffmann C, Dieterle M, Moreau F, Neumann K, Papuga J, Furtado AT, Steinmetz A, Thomas C (2015) The pH sensibility of actin-bundling LIM proteins is governed by the acidic properties of their C-terminal domain. FEBS Lett 589:2312–2319
Mundel C, Baltz R, Eliasson Å, Bronner R, Grass N, Kräuter R, Evrard JL, Steinmetz A (2000) A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol Biol 42:291–302
Na JK, Huh SM, Yoon IS, Byun MO, Lee YH, Lee KO, Kim DY (2014) Rice LIM protein OsPLIM2a is involved in rice seed and tiller development. Mol Breed 34:569–581
Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A, Thomas C (2010) Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22:3034–3052
Park JI, Ahmed NU, Jung HJ, Arasan SK, Chung MY, Cho YG, Watanabe M, Nou IS (2014) Identification and characterization of LIM gene family in Brassica rapa. BMC Genom 15:641
Peng Y, Ma W, Chen L, Yang L, Li S, Zhao H, Zhao Y, Jin W, Li N, Bevan MW, Li X, Tong Y, Li Y (2013) Control of root meristem size by DA1-RELATED PROTEIN2 in Arabidopsis. Plant Physiol 161:1542–1556
Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 273:649–662
Pérez-Alvarado GC, Miles C, Michelsen JW, Louis HA, Winge DR, Beckerle MC, Summers MF (1994) Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Mol Biol 1:388–398
Porter K, Day B (2016) From filaments to function: the role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J Integr Plant Biol 58: 299–311
Sadler I, Crawford AW, Michelsen JW, Beckerle MC (1992) Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol 119:1573–1587
Sánchez-García I, Rabbits TH (1994) The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet 10:315–320
Schmeichel KL, Beckerle MC (1997) Molecular dissection of a LIM domain. Mol Biol Cell 8:219–230
Skau CT, Courson DS, Bestul AJ, Winkelman JD, Rock RS, Sirotkin V, Kovar DR (2011) Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J Biol Chem 286:26964–26977
Srivastava V, Verma PV (2015) Genome wide identification of LIM genes in Cicer arietinum and response of Ca-2LIMs in development, hormone and pathogenic stress. PLoS One 10:e0138719
Sweetman J, Spurr C, Eliasson A, Gass N, Steinmetz A, Twell D (2000) Isolation and characterisation of two pollen-specific LIM domain protein cDNAs from Nicotiana tabacum. Sex Plant Reprod 12(6):339–345
Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, Steinmetz A (2006) Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18:2194–2206
Thomas C, Moreau F, Dieterle M, Hoffmann C, Gatti S, Hofmann C, Van Troys M, Ampe C, Steinmetz A (2007a) The LIM domains of WLIM1 define a new class of actin bundling modules. J Biol Chem 282:33599–33608
Thomas C, Hoffmann C, Gatti S, Steinmetz A (2007b) LIM proteins: a novel class of actin cytoskeleton organizers in plants. Plant Signal Behav 2:99–100
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20
Wang HJ, Wan AR, Jauh GY (2008) An actin binding protein, LlLIM1, mediates Ca2+ and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol 147:1619–1636
Wang X, Liu B, Huang C, Zhang X, Luo C, Cheng X, Yu R, Wu Z (2012) Over expression of Zmda1-1 gene increases seed mass of corn. Afr J Biotechnol 11(69):13387–13395
Way JC, Chalfie M (1988) Mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54:5–16
Weiskirchen R, Günther K (2003) The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays 25:152–162
Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25:3347–3359
Xu F, Zhu C, Cevik V, Johnson K, Liu Y, Sohn K, Jones JD, Holub EB, Li X (2015) Autoimmunity conferred by chs3-2D relies on CSA1, its adjacent TNL-encoding neighbour. Sci Rep 5:8792
Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S (2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63:283–296
Yao X, Pérez-Alvarado GC, Louis HA, Pomiès P, Hatt C, Summers MF, Beckerle MC (1999) Solution structure of the chicken cysteine-rich protein, CRP1, a double-LIM protein implicated in muscle differentiation. Biochem 38:5701–5713
Ye J, Xu M (2012) Actin bundler PLIM2s are involved in the regulation of pollen development and tube growth in Arabidopsis. J Plant Physiol 169:516–522
Zhang X, Liu CJ (2015) Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant 8:17–27
Zhao M, He L, Gu Y, Wang Y, Chen Q, He C (2014) Genome-wide analyses of a plant-specific LIM-domain gene family implicate its evolutionary role in plant diversification. Genom Biol Evol 6:1000–1012
Zhao M, Gu Y, He L, Chen Q, He C (2015) Sequence and expression variations suggest an adaptive role for the DA1-like gene family in the evolution of soybeans. BMC Plant Biol 15:120
Zheng Q, Zhao Y (2007) The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein–protein interaction. Bio Cell 99:489–502
Acknowledgements
Authors acknowledge Central University of Jammu (CUJ), Jammu and National Institute of Plant Genome Research (NIPGR), New Delhi for work facility. This work is supported by the Department of Biotechnology project, Government of India (BT/AGR/CG-Phase II/01/2014) and Council of Scientific and Industrial Research (CSIR), India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Srivastava, V., Verma, P.K. The plant LIM proteins: unlocking the hidden attractions. Planta 246, 365–375 (2017). https://doi.org/10.1007/s00425-017-2715-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2715-7