Abstract
Main Conclusion
In the present study, miRNA precursors in the genomes of three palm species were identified. Analyzes of sequence conservation and biological function of their putative targets contribute to understand the roles of miRNA in palm biology.
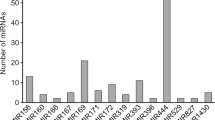
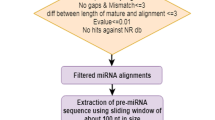
MicroRNAs are small RNAs of 20–25 nucleotides in length, with important functions in the regulation of gene expression. Recent genome sequencing of the palm species Elaeis guineensis, Elaeis oleifera and Phoenix dactylifera have enabled the discovery of miRNA genes, which can be used as biotechnological tools in palm trees breeding. The goal of this study is the identification of miRNA precursors in the genomes of these species and their possible biological roles suggested by the mature miRNA-based regulation of target genes. Mature miRNA sequences from Arabidopsis thaliana, Oryza sativa, and Zea mays available at the miRBase were used to predict microRNA precursors in the palm genomes. Three hundred and thirty-eight precursors, ranging from 76 to 220 nucleotide (nt) in size and distributed in 33 families were identified. Moreover, we also identified 266 miRNA precursors of Musa acuminata, which are phylogenetically close to palms species. To understand the biological function of palm miRNAs, 374 putative miRNA targets were identified. An enrichment analysis of target-gene function was carried out using the agriGO tool. The results showed that the targets are involved in plant developmental processes, mainly regulating root development. Our findings contribute to increase the knowledge on microRNA roles in palm biology and could help breeding programs of palm trees.






Similar content being viewed by others
Abbreviations
- CDS:
-
Coding DNA sequence
- EST:
-
Expressed sequence tag
- GO:
-
Gene ontology
References
Akinoso R, Raji AO (2011) Physical properties of fruit, nut and kernel of oil palm. Int Agrophysics 25:85–88
Al-Mssallem IS, Hu S, Zhang X et al (2013) Genome sequence of the date palm Phoenix dactylifera L. Nat Commun 4:2274. doi:10.1038/ncomms3274
Al-Shahib W, Marshall RJ (2003) The fruit of the date palm: its possible use as the best food for the future? Int J Food Sci Nutr 54:247–259
Barcelos E, Rios SDA, Cunha RNV et al (2015) Oil palm natural diversity and the potential for yield improvement. Front Plant Sci 6:190. doi:10.3389/fpls.2015.00190
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116:281–297
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933. doi:10.1073/pnas.0505461102
Bologna NG, Schapire AL, Palatnik JF (2013) Processing of plant microRNA precursors. Brief Funct Genomics 12:37–45. doi:10.1093/bfgp/els050
Breakfield NW, Corcoran DL, Petricka JJ et al (2011) High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res 22:163–176. doi:10.1101/gr.123547.111
Camillo J, Braga VC, Mattos JKDA et al (2014) Seed biometric parameters in oil palm accessions from a Brazilian germplasm bank. Pesqui Agropecuária Bras 49:604–612. doi:10.1590/S0100-204X2014000800004
Cao X, Bourgis F, Kilaru A et al (2011) Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA 108:12527–12532. doi:10.1073/pnas.1115243108
Chai J, Feng R, Shi H et al (2015) Bioinformatic identification and expression analysis of banana microRNAs and their targets. PLoS ONE 10:e0123083. doi:10.1371/journal.pone.0123083
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896. doi:10.1038/nrg2179
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. doi:10.1146/annurev.cellbio.042308.113417
Cochard B, Amblard P, Durand-Gasselin T (2005) Oil palm genetic improvement and sustainable development. Oléagineux Corps gras Lipides 12:141–147
Comai L, Zhang B (2012) MicroRNAs: key gene regulators with versatile functions. Plant Mol Biol 80:1
Corley RHV, Tinker PB (2003) The palm oil, 4th edn. Blackwell, Oxford
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442. doi:10.1105/tpc.110.082784
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39. Suppl 2:W155–W159. doi:10.1093/nar/gkr319
Dezulian T, Palatnik JF, Huson D, Weigel D (2005) Conservation and divergence of microRNA families in plants. Genome Biol 6:P13
Ding D, Li W, Han M et al (2014) Identification and characterisation of maize microRNAs involved in developing ears. Plant Biol 16:9–15. doi:10.1111/plb.12013
Eiserhardt WL, Svenning JC, Kissling WD, Balslev H (2011) Geographical ecology of the palms (Arecaceae): determinants of diversity and distributions across spatial scales. Ann Bot 108:1391–1416. doi:10.1093/aob/mcr146
Ghag SB, Shekhawat UKS, Ganapathi TR (2015) Small RNA profiling of two important cultivars of banana and overexpression of miRNA156 in transgenic banana plants. PLoS One 10:e0127179. doi:10.1371/journal.pone.0127179
Gorodkin J, Havgaard JH, Ensterö M et al (2006) MicroRNA sequence motifs reveal asymmetry between the stem arms. Comput Biol Chem 30:249–254. doi:10.1016/j.compbiolchem.2006.04.006
Griffiths-Jones S, Grocock RJ, Van Dongen S et al (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Hsieh L-C, Lin S-I, Shih AC-C et al (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132. doi:10.1104/pp.109.147280
Ithnin M, Singh R, Din AK (2011) Elaeis. In: Kole C (ed) Wild crop relatives: Genomic and breeding resources. Plantation and ornamental crops. Springer, Berlin Heidelberg, pp 113–124
Jain SM, Al-Khayri JM, Johnson DV (2011) Date palm biotechnology, 1st edn. Springer, Netherlands. doi:10.1007/978-94-007-1318-5
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Katiyar A, Smita S, Muthusamy S et al (2015) Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front. Plant Sci 6:506. doi:10.3389/fpls.2015.00506
Khan GA, Declerck M, Sorin C et al (2011) MicroRNAs as regulators of root development and architecture. Plant Mol Biol 77:47–58. doi:10.1007/s11103-011-9793-x
Khraiwesh B, Zhu J-K, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148. doi:10.1016/j.bbagrm.2011.05.001
Kidner CA (2010) The many roles of small RNAs in leaf development. J Genet Genom 37:13–21. doi:10.1016/S1673-8527(09)60021-7
Kulcheski FR, de Oliveira LF, Molina LG et al (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom 12:307. doi:10.1186/1471-2164-12-307
Lacombe S, Nagasaki H, Santi C et al (2008) Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Biol 8:123. doi:10.1186/1471-2229-8-123
Lakhotia N, Joshi G, Bhardwaj AR et al (2014) Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol 14:6. doi:10.1186/1471-2229-14-6
Lee WS, Gudimella R, Wong GR et al (2015) Transcripts and microRNAs responding to salt stress in Musa acuminata Colla (AAA Group) cv. Berangan Roots. PLoS One 10:e0127526. doi:10.1371/journal.pone.0127526
Low ETL, Rosli R, Jayanthi N et al (2014) Analyses of hypomethylated oil palm gene space. PLoS One 9:e86728. doi:10.1371/journal.pone.0086728
Ma Z, Coruh C, Axtell MJ (2010) Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22:1090–10103. doi:10.1105/tpc.110.073882
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38(Suppl):S31–S36. doi:10.1038/ng1791
Meerow AW, Krueger RR, Singh R, et al. (2012) Coconut, date, and oil palm genomics. In: Schnell RJ, Priyadarshan PM (eds) Genomics of tree crops. Springer, New York, pp 299–351. doi:10.1007/978-1-4614-0920-5_10
Mehrpooyan F, Othman RY, Harikrishna JA (2012) Tissue and temporal expression of miR172 paralogs and the AP2-like target in oil palm (Elaeis guineensis Jacq.). Tree Genet Genom 8:1331–1343
Meng Y, Shao C, Wang H, Chen M (2011) The regulatory activities of plant microRNAs: a more dynamic perspective. Plant Physiol 157:1583–1595. doi:10.1104/pp.111.187088
Meyers BC, Souret FF, Lu C et al (2006) Sweating the small stuff: microRNA discovery in plants. Curr Opin Biotechnol 17:139–146. doi:10.1016/j.copbio.2006.01.008
Mica E, Piccolo V, Delledonne M et al (2010) Correction: high throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genom 11:109. doi:10.1186/1471-2164-11-109
Mishra NS, Mukherjee SK (2007) A peep into the plant miRNA world. Open Plant Sci J 1:1–9. doi:10.2174/187429470701011074
Nasaruddin N, Harikrishna K, Othman RY et al (2007) Computational prediction of microRNAs from oil palm (Elaeis guineensis Jacq.) expressed sequence tags. AsPac J Mol Biol Biotechnol 15:107–113
Reinhart BJ, Weinstein EG, Rhoades MW et al (2002) MicroRNAs in plants. Genes Dev 16:1616–1626. doi:10.1101/gad.1004402
Silva GFFE, Silva EM, Da Silva MA et al (2014) MicroRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78:604–618. doi:10.1111/tpj.12493
Singh R, Low E-TL, Ooi LC-L et al (2013a) The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 500:340–344. doi:10.1038/nature12356
Singh R, Ong-Abdullah M, Low E-TL et al (2013b) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500:335–339. doi:10.1038/nature12309
Souret F, Lu C, Green PJ, Meyers BC (2005) Sweating the small stuff: microRNA discovery in plants. Curr Opin Biotechnol 17:139–146. doi:10.1016/j.copbio.2006.01.008
Sun G (2012) MicroRNAs and their diverse functions in plants. Plant Mol Biol 80:17–36. doi:10.1007/s11103-011-9817-6
Thiebaut F, Grativol C, Carnavale-bottino M et al (2012) Computational identification and analysis of novel sugarcane microRNAs. BMC Genom 13:290. doi:10.1186/1471-2164-13-290
Thiebaut F, Grativol C, Tanurdzic M et al (2014) Differential sRNA regulation in leaves and roots of sugarcane under water depletion. PLoS ONE 9:4. doi:10.1371/journal.pone.0093822
Wang X-J, Reyes JL, Chua N-H, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5:R65. doi:10.1186/gb-2004-5-9-r65
Wu L, Zhou H, Zhang Q et al (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38:465–475. doi:10.1016/j.molcel.2010.03.008
Xiao Y, Xia W, Yang Y et al (2013) Characterization and evolution of conserved microRNA through duplication events in date palm (Phoenix dactylifera). PLoS One 8:8. doi:10.1371/journal.pone.0071435
Xin C, Liu W, Lin Q et al (2015) Profiling microRNA expression during multi-staged date palm (Phoenix dactylifera L.) fruit development. Genomics 105:242–251. doi:10.1016/j.ygeno.2015.01.004
Xu X, Jiang Q, Ma X et al (2014) Deep sequencing identifies tissue-specific microRNAs and their target genes involving in the biosynthesis of tanshinones in Salvia miltiorrhiza. PLoS One 9:e111679. doi:10.1371/journal.pone.0111679
Zhang BH, Pan XP, Wang QL et al (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360. doi:10.1038/sj.cr.7290302
Zhang B, Pan X, Anderson TA (2006a) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762. doi:10.1016/j.febslet.2006.05.063
Zhang B, Pan X, Cannon CH et al (2006b) Conservation and divergence of plant microRNA genes. Plant J 46:243–259. doi:10.1111/j.1365-313X.2006.02697.x
Zhang B, Pan X, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182. doi:10.1007/s00425-008-0818-x
Zhang J, Xu Y, Huan Q, Chong K (2009) Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genom 10:449
Zhang X, Zhao H, Gao S et al (2011) Arabidopsis Argonaute 2 regulates innate immunity via miR393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42:356–366. doi:10.1016/j.molcel.2011.04.010
Zhang F, Dong W, Huang L et al (2015) Identification of microRNAs and their targets associated with embryo abortion during chrysanthemum cross breeding via high-throughput sequencing. PLoS One 10:e0124371. doi:10.1371/journal.pone.0124371
Acknowledgments
The authors thank FAPERJ (Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for the financial support. Authors are grateful to Sergei Kouchnir and Edson Barcelos da Silva for providing plants pictures. PCGF, ASH are grateful to the CNPq and FAPERJ, CG and FT are grateful to FAPERJ, and ACS is grateful to FAPESPA (Fundação Amazônica de Amparo a Estudos e Pesquisas do Pará) for fellowships.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
da Silva, A.C., Grativol, C., Thiebaut, F. et al. Computational identification and comparative analysis of miRNA precursors in three palm species. Planta 243, 1265–1277 (2016). https://doi.org/10.1007/s00425-016-2486-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2486-6