Abstract
Main conclusion
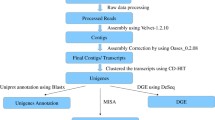
The transcriptomes of Aconitum heterophyllum were assembled and characterized for the first time to decipher molecular components contributing to biosynthesis and accumulation of metabolites in tuberous roots.
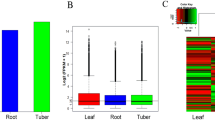
Aconitum heterophyllum Wall., popularly known as Atis, is a high-value medicinal herb of North-Western Himalayas. No information exists as of today on genetic factors contributing to the biosynthesis of secondary metabolites accumulating in tuberous roots, thereby, limiting genetic interventions towards genetic improvement of A. heterophyllum. Illumina paired-end sequencing followed by de novo assembly yielded 75,548 transcripts for root transcriptome and 39,100 transcripts for shoot transcriptome with minimum length of 200 bp. Biological role analysis of root versus shoot transcriptomes assigned 27,596 and 16,604 root transcripts; 12,340 and 9398 shoot transcripts into gene ontology and clusters of orthologous group, respectively. KEGG pathway mapping assigned 37 and 31 transcripts onto starch–sucrose metabolism while 329 and 341 KEGG orthologies associated with transcripts were found to be involved in biosynthesis of various secondary metabolites for root and shoot transcriptomes, respectively. In silico expression profiling of the mevalonate/2-C-methyl-d-erythritol 4-phosphate (non-mevalonate) pathway genes for aconites biosynthesis revealed 4 genes HMGR (3-hydroxy-3-methylglutaryl-CoA reductase), MVK (mevalonate kinase), MVDD (mevalonate diphosphate decarboxylase) and HDS (1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase) with higher expression in root transcriptome compared to shoot transcriptome suggesting their key role in biosynthesis of aconite alkaloids. Five genes, GMPase (geranyl diphosphate mannose pyrophosphorylase), SHAGGY, RBX1 (RING-box protein 1), SRF receptor kinases and β-amylase, implicated in tuberous root formation in other plant species showed higher levels of expression in tuberous roots compared to shoots. A total of 15,487 transcription factors belonging to bHLH, MYB, bZIP families and 399 ABC transporters which regulate biosynthesis and accumulation of bioactive compounds were identified in root and shoot transcriptomes. The expression of 5 ABC transporters involved in tuberous root development was validated by quantitative PCR analysis. Network connectivity diagrams were drawn for starch–sucrose metabolism and isoquinoline alkaloid biosynthesis associated with tuberous root growth and secondary metabolism, respectively, in root transcriptome of A. heterophyllum. The current endeavor will be of practical importance in planning a suitable genetic intervention strategy for the improvement of A. heterophyllum.











Similar content being viewed by others
Abbreviations
- AHSR:
-
Root transcriptome
- AHSS:
-
Shoot transcriptome
- COG:
-
Cluster of orthologous group
- GO:
-
Gene ontology
- KO:
-
KEGG orthology
- MEP:
-
2-C-Methyl-d-erythritol 4-phosphate
- MVA:
-
Mevalonate
- NGS:
-
Next-generation sequencing
- FPKM:
-
Fragments per kilobase of transcript per million fragments mapped
- SSR:
-
Simple sequence repeat
References
Bhattacharyya D, Sinha R, Hazra S, Datta R, Chattopadhyay S (2013) De novo transcriptome analysis using 454 pyrosequencing of the Himalayan Mayapple, Podophyllum hexandrum. BMC Genomics 14:748
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96:4198–4203
Costa GGL, Cardoso KC, Del Bem LEV, Lima AC, Cunha MAS, de Campos-Leite L, Vicentini R, Papes F, Moreira RC, Yunes JA, Campos FAP, Da Silva MJ (2010) Transcriptome analysis of the oil-rich seed of the bioenergy crop Jatropha curcas L. BMC Genom 11:462
Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot 99:787–822
Endress PK (2002) Morphology and angiosperm systematics in the molecular era. Bot Rev 68:545–570
Gahlan P, Singh RH, Shankar R, Sharma N, Kumari A, Chawla V, Ahuja PS, Kumar S (2012) De novo sequencing and characterization of Picrorhiza kurrooa transcriptome at two temperatures showed major transcriptome adjustments. BMC Genom 13:126
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63
Geer LY, Bauer-Marchler A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH (2009) The NCBI biosystems database. Nucleic Acids Res 38:492–496
Ginis O, Courdavault V, Melin C, Lanoue A, Giglioli-Guivarc’h N, St-Pierre B, Courtois M, Oudin A (2012) Molecular cloning and functional characterization of Catharanthus roseus hydroxymethylbutenyl 4-diphosphate synthase gene promoter from the methyl erythritol phosphate pathway. Mol Biol Rep 39:5433–5447
Goff S, Ricke D, Lan T, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Gupta N, Sharma SK, Rana JC, Chauhan RS (2011) Expression of flavonoid biosynthesis genes vis-à-vis rutin content variation in different growth stages of Fagopyrum species. J Plant Physiol 168:2117–2123
Hudson HE (2008) Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol Ecol Resour 8:3–17
Hussain MS, Fareed S, Saba Ansari M, Rahman A, Ahmad IZ, Saeed M (2012) Current approaches toward production of secondary plant metabolites. J Pharmacy Bioallied Sci 4:10–20
Ideker T, Krogen NJ (2012) Differential network biology. Mol Syst Biol 8:565
Ihemere U, Arias-Garzon D, Lawrence S, Sayre R (2006) Genetic modification of cassava for enhanced starch production. Plant Biotechnol J 4:453–465
Indira P, Kurian T (1977) A study on the comparative anatomical changes undergoing tuberization in the roots of cassava and sweet potato. J Root Crops 3:29–32
Initiative TAG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Jaiswal V, Chanumolu SK, Gupta A, Chauhan RS, Rout C (2013) Jenner-predict server: prediction of protein vaccine candidates (PVCs) in bacteria based on host-pathogen interactions. BMC Bioinformatics 14:211
Jin J, Zhang H, Kong L, Gao G, Luo J (2013) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid insensitive locus in Arabidopsis. Plant Physiol 127:14–22
Li C, Li QG, Dunwell JM, Zhang YM (2012a) Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol Biol Evol 29:3227–3236
Li DJ, Xia ZH, Deng Z, Liu XH, Dong JM, Feng FY (2012b) Development and characterization of intron-flanking EST-PCR markers in rubber tree (Hevea brasiliensis Muell. Arg.). Mol Biotechnol 51:148–159
Liu T, Zhu S, Tang Q, Chen P, Yu Y, Tang S (2013) De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genom 14:125
Malhotra N, Kumar V, Sood H, Singh TR, Chauhan RS (2014) Multiple genes of mevalonate and non-mevalonate pathways contribute to high aconites content in an endangered medicinal herb, Aconitum heterophyllum Wall. Phytochemistry 108:26–34
Memelink J, Verpoorte R, Kijne JW (2001) ORCAnisation of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci 6:212–219
Mitra SK, Ashish S, Udupa V, Jayakumar K (2001) Experimental evaluation of diarex vet in lactose induced diarrhoea in rats. Indian Vet J 78:212–216
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Nautiyal BP, Prakash V, Bahuguna R, Maithani UC, Bisht H, Nautiyal MC (2002) Population study for monitoring the status of rarity of three Aconite species in Garhwal Himalaya. Tropical Ecol 43:297–303
Nogués I, Brilli F, Loreto F (2006) Dimethylallyl diphosphate and geranyl diphosphate pools of plant species characterized by different isoprenoid emissions. Plant Physiol 141:721–730
Pelletier SW, Aneja R, Gopinath KW (1968) The alkaloids of Aconitum heterophyllum Wall: isolation and characterization. Phytochemistry 7:625–635
Rico C, Normandeau E, Dion-Côté AM, Rico MI, Côté G, Bernatchez L (2013) Combining next-generation sequencing and online databases for microsatellite development in non-model organisms. Sci Rep 3:3376
Saithong T, Rongsirikul O, Kalapanulak S, Chiewchankaset P, Siriwat W, Netrphan S, Suksangpanomrung M, Meechai A, Cheevadhanarak S (2013) Starch biosynthesis in cassava: a genome-based pathway reconstruction and its exploitation in data integration. BMC Syst Biol 7:75
Schnable P, Ware D, Fulton R, Stein J, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves T (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schulte AE, Llamas Durán AM, van der Heijden R, Verpoorte R (2000) Mevalonate kinase activity in Catharanthus roseus plants and suspension cultured cells. Plant Sci 150:59–69
Sheffield J, Taylor N, Fauquet C, Chen S (2006) The cassava (Manihot esculenta Crantz) root proteome: protein identification and differential expression. Proteomics 6:1588–1598
Sigurdsson MI, Jamshidi N, Steingrimsson E, Thiele I, Palsson BA (2010) A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst Biol 4:140
Singh RS, Gara RK, Bhardwaj PK, Kaachra A, Malik S, Kumar R, Sharma M, Ahuja PS, Kumar S (2010) Expression of 3-hydroxy-3-methylglutaryl-CoA reductase, p-hydroxybenzoate-m-geranyltransferase and genes of phenylpropanoid pathway exhibits positive correlation with shikonins content in arnebia [Arnebia euchroma (Royle) Johnston]. BMC Mol Biol 11:88
Srivastava N, Sharma V, Dobriyal AK, Kamal B, Gupta S, Jadon VS (2011) Influence of pre-sowing treatments on in vitro seed germination of Ativisha (Aconitum heterophyllum Wall.) of Uttarakhand. Biotechnology 10:215–219
Sun P, Guo Y, Qi J, Zhou L, Li X (2010) Isolation and expression analysis of tuberous root development related genes in Rehmannia glutinosa. Mol Biol Rep 37:1069–1079
Tanaka M, Takahata Y, Nakatani M (2005) Analysis of genes developmentally regulated during storage root formation of sweet potato. J Plant Physiol 162:91–102
Thatte UM, Rege NN, Phatak SD, Dahanukar SA (1993) The flip side of ayurveda. J Postgrad Med 39:179–182
Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the differences in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104:1–8
Wang QM, Zhang LM, Wang ZL (2005) Formation and thickening of tuberous roots in relation to the endogenous hormone concentrations in sweetpotato. Scientia Agricultura Sinica 38:2414–2420
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang B, Guo G, Wang C, Lin Y, Wang X, Zhao M, Guo Y, He M, Zhang Y, Pan L (2010a) Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res 38:5075–5087
Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, Liu SS (2010b) De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genom 11:400
Wang Z, Fang B, Chen J, Zhang X, Luo Z, Huang L, Chen X, Li Y (2010c) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas). BMC Genom 11:726
Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Lv H, Zhang X (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genom 12:451
Xia ZH, Xu HM, Zhai JL, Li DJ, He CZ, Huang X (2011) RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol Biol 77:299–308
Xie W, Lei Y, Fu W, Yang Z, Zhu X, Guo Z, Wu Q, Wang S, Xu B, Zhou X, Zhang Y (2012) Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut. Int J Biol Sci 8:1142–1155
Yazaki K (2005) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580:1183–1191
You MK, Hur CG, Ahn YS, Suh MC, Jeong BC, Shin JS, Bae JM (2003) Identification of genes possibly related to storage root induction in sweet potato. FEBS Lett 536:101–105
Zhaohong W, Wang J, Xing J, He Y (2006) Quantitative determination of alkaloids in four species of Aconitum by HPLC. J Pharma Biomed Anal 40:1031–1034
Acknowledgments
The authors are thankful to the Department of Biotechnology, Ministry of Science and Technology, Government of India, for providing funds to RSC in the form of a program support on high-value medicinal plants. The authentic plant material provided by Dr. Sandeep Sharma, HFRI, Panthaghati, Shimla (HP), is also acknowledged.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Tarun Pal and Nikhil Malhotra contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pal, T., Malhotra, N., Chanumolu, S.K. et al. Next-generation sequencing (NGS) transcriptomes reveal association of multiple genes and pathways contributing to secondary metabolites accumulation in tuberous roots of Aconitum heterophyllum Wall.. Planta 242, 239–258 (2015). https://doi.org/10.1007/s00425-015-2304-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2304-6