Abstract
Main conclusion
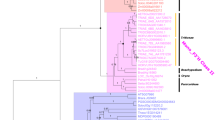
Multiple F3′5′H evolution from F3′H has occurred in dicotyledonous plants. Efficient pollinator attraction is probably the driving force behind, as this allowed for the synthesis of delphinidin-based blue anthocyanins.
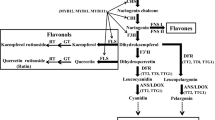
The cytochrome P450-dependent monooxygenases flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) hydroxylate the B-ring of flavonoids at the 3′- and 3′- and 5′-position, respectively. Their divergence took place early in plant evolution. While F3′H is ubiquitously present in higher plants, the distribution of F3′5′H is scattered. Here, we report that F3′5′H has repeatedly evolved from F3′H precursors at least four times in dicotyledonous plants: In the Asteraceae, we identified F3′5′Hs specific for the subfamilies Cichorioideae and Asteroideae, and additionally an F3′5′H that seems to be specific for the genus Echinops of the subfamily Carduoideae; moreover, characterisation of a sequence from Billardiera heterophylla (formerly Sollya heterophylla) (Pittosporaceae) showed that the independent evolution of an F3′5′H has occurred at least once also in another family. The evolution of F3′5′H from an F3′H precursor represents a gain of enzymatic function, probably triggered by an amino acid change at one position of substrate recognition site 6. The gain of F3′5′H activity allows for the synthesis of delphinidin-based anthocyanins which usually provide the basis for lilac to blue flower colours. Therefore, the need for an efficient pollinator attraction is probably the driving force behind the multiple F3′5′H evolution.




Similar content being viewed by others
Abbreviations
- BEBm:
-
Bayes empirical Bayes
- Cy:
-
Cyanidin
- CYP:
-
Cytochrome P450
- Dp:
-
Delphinidin
- GOF:
-
Gain of function
- LOF:
-
Loss of function
- LRT:
-
Likelihood ratio test
- MSA:
-
Multiple sequence alignment
- Pg:
-
Pelargonidin
- SRS:
-
Substrate recognition site
References
Baudry J, Li W, Pan L, Berenbaum MR, Schuler MA (2003) Molecular docking of substrates and inhibitors in the catalytic site of CYP6B1, an insect cytochrome P450 monooxygenase. Protein Eng 16:577–587
Blackburne BP, Whelan S (2012) Class of multiple sequence alignment algorithm affects genomic analysis. Mol Biol Evol 30:256
Boase M, Lewis D, Davies K, Marshall G, Patel D, Schwinn K, Deroles S (2010) Isolation and antisense suppression of flavonoid 3′, 5′-hydroxylase modifies flower pigments and colour in cyclamen. BMC Plant Biol 10:107
Bohm BA, Stuessy TF (2001) Flavonoids of the sunflower family (Asteraceae). Springer Verlag, Wien
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bremer K, Major (1994) Clades and grades of the Asteraceae. In: Hind DJN BH (ed) Compositae: systematics. Proceedings of the International Compositae Conference, Kew, pp 1–7
Brouillard R (1983) The in vivo expression of anthocyanin colour in plants. Phytochemistry 22:1311–1323
Brugliera F, Demelis L, Koes R, Tanaka Y (2003) Genetically modified plants with altered inflorescence. BRUGLIERA, Filippa. US Patent Nr. 8,288,612, 2012
Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52:98–111
Chen JS, Berenbaum M, Schuler M (2002) Amino acids in SRS1 and SRS6 are critical for furanocoumarin metabolism by CYP6B1v1, a cytochrome P450 monooxygenase. Insect Mol Biol 11:175–186
Cronk Q, Ojeda I (2008) Bird-pollinated flowers in an evolutionary and molecular context. J Exp Bot 59:715–727
Des Marais DL, Rausher MD (2008) Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454:762–765
Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids–a gold mine for metabolic engineering. Trends Plant Sci 4:394–400
Fletcher W, Yang Z (2010) The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27:2257–2267
Forkmann G, Heller W (1999) Biosynthesis of flavonoids. In: Barton D, Nakanishi K, Meth-Cohn O, Sankawa U (eds) Comprehensive natural products chemistry, vol 1. Elsevier Science, Amsterdam, pp 713–748
Goertzen LR, Cannone JJ, Gutell RR, Jansen RK (2003) ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Mol Phylogenet Evol 29:216–234
Gotoh O (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Halbwirth H, Muster G, Stich K (2008) Unraveling the biochemical base of Dahlia flower coloration. Nat Prod Comm 3:1259–1266
Harborne JB (1967) Comparative biochemistry of flavonoids. Academic Press, London
Harborne JB (1993) Introduction to ecological biochemistry. Academic Press, London
Harborne JB, Baxter H (1999) The handbook of natural flavonoids, Vol. 1 and Vol. 2. Wiley, New York
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3:41–62
Ishiguro K, Taniguchi M, Tanaka Y (2012) Functional analysis of Antirrhinum kelloggii flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes; critical role in flower color and evolution in the genus Antirrhinum. J Plant Res 125:451–456
Jansen R, Kim KJ (1994) Implications of chloroplast DNA data for the classification and phylogeny of the Asteraceae. In: Hind DJN BH (ed) Compositae: systematics. Proceedings of the International Compositae Conference, Kew, pp 317–339
Jordan G, Goldman N (2011) The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol Biol Evol 29:272
Kaltenbach M, Schröder G, Schmelzer E, Lutz V, Schröder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19:183–193
Katoh K, Misawa K, Ki Kuma, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Löytynoja A, Goldman N (2005) An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA 102:10557–10562
Markham K (1982) Techniques of flavonoid identification. Academic Press, London
Martens S, Knott J, Seitz CA, Janvari L, Yu SN, Forkmann G (2003) Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissues. Biochem Eng J 14:227–235
Matern U, Reichenbach C, Heller W (1986) Efficient uptake of flavonoids into parsley (Petroselinum hortense) vacuoles requires acylated glycosides. Planta 167:183–189
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nelson D, Werck-Reichhart D (2011) A P450-centric view of plant evolution. Plant J 66:194–211
Nørbæk R, Nielsen K, Kondo T (2002) Anthocyanins from flowers of Cichorium intybus. Phytochemistry 60:357–359
Nordstrom C, Swain T (1953) The flavonoid glycosides of Dahlia variabilis. J Chem Soc 555:2764
Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450 s in optimized redox environments. Methods Enzymol 272:51–64
Rausher MD (2008) Evolutionary transitions in floral color. Int J Plant Sci 169:7–21
Redelings B (2014) Erasing errors due to alignment ambiguity when estimating positive selection. Mol Biol Evol 31:174
Rupasinghe S, Baudry J, Schuler MA (2003) Common active site architecture and binding strategy of four phenylpropanoid P450 s from Arabidopsis thaliana as revealed by molecular modeling. Protein Eng 16:721–731
Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54:629–667
Schlangen K, Miosic S, Topuz F, Muster G, Marosits T, Seitz C, Halbwirth H (2009) Chalcone 3-hydroxylation is not a general property of flavonoid 3′-hydroxylase. Plant Sci 177:97–102
Schlangen K, Miosic S, Halbwirth H (2010) Allelic variants from Dahlia variabilis encode flavonoid 3′-hydroxylases with functional differences in chalcone 3-hydroxylase activity. Arch Biochem Biophys 494:40–45
Schwinn K, Miosic S, Davies K, Thill J, Gotame TP, Stich K, Halbwirth H (2014) The B-ring hydroxylation pattern of anthocyanins can be determined through activity of the flavonoid 3′-hydroxylase on leucoanthocyanidins. Planta 240:1003–1010
Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G (2006) Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′, 5′-hydroxylase in the Asteraceae family. Plant Mol Biol 61:365–381
Seitz C, Ameres S, Forkmann G (2007a) Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase. FEBS Lett 581:3429–3434
Seitz C, Vitten M, Steinbach P, Hartl S, Hirsche J, Rathje W, Treutter D, Forkmann G (2007b) Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry 68:824–833
Stich K, Eidenberger T, Wurst F, Forkmann G (1992) Enzymatic conversion of dihydroflavonols to flavan-3, 4-diols using flower extracts of Dianthus caryophyllus L. (carnation). Planta 187:103–108
Streisfeld MA, Rausher MD (2009) Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. New Phytol 183:751–763
Suchard MA, Redelings BD (2006) BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics 22:2047–2048
Takeda K, Osakabe A, Saito S, Furuyama D, Tomita A, Kojima Y, Yamadera M, Sakuta M (2005) Components of protocyanin, a blue pigment from the blue flowers of Centaurea cyanus. Phytochemistry 66:1607–1613
Tanaka Y, Brugliera F, Chandler S (2009) Recent progress of flower colour modification by biotechnology. Int J Mol Sci 10:5350–5369
Tanaka Y, Brugliera F, Kalc G, Senior M, Dyson B, Nakamura N, Katsumoto Y, Chandler S (2010) Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosc Biotech Biochem 74:1760–1769
Wagner A (2002) Selection and gene duplication: a view from the genome. Genome Biol 3:1012
WenHsiung L (1997) Molecular evolution. Sinauer associates incorporated, Sunderland
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917
Yang Z, Wong WS, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Yoshida K, Kondo T (2009) Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat Prod Rep 26:884–915
Zufall RA, Rausher MD (2004) Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature 428(6985):847–850
Acknowledgments
H. Halbwirth acknowledges funding by the Austrian Science Fund FWF (P24331-B16).
Author information
Authors and Affiliations
Corresponding author
Additional information
Special topic: Polyphenols: biosynthesis and function in plants and ecosystems. Guest editor: Stefan Martens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seitz, C., Ameres, S., Schlangen, K. et al. Multiple evolution of flavonoid 3′,5′-hydroxylase. Planta 242, 561–573 (2015). https://doi.org/10.1007/s00425-015-2293-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2293-5