Abstract
Main conclusion
In contrast to current knowledge, the B -ring hydroxylation pattern of anthocyanins can be determined by the hydroxylation of leucoanthocyanidins in the 3′ position by flavonoid 3’-hydroxylase.
Abstract
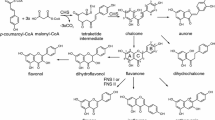
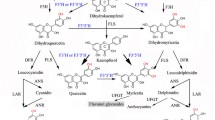
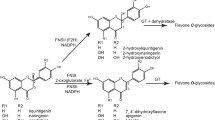
The cytochrome P450-dependent monooxygenases flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) are key flavonoid enzymes that introduce B-ring hydroxyl groups in positions 3′ or 3′ and 5′, respectively. The degree of B-ring hydroxylation is the major determinant of the hue of anthocyanin pigments. Numerous studies have shown that F3′H and F3′5′H may act on more than one type of anthocyanin precursor in addition to other flavonoids, but it has been unclear whether the anthocyanin precursor of the leucoanthocyanidin type can be hydroxylated as well. We have investigated this in vivo using feeding experiments and in vitro by studies with recombinant F3′H. Feeding leucoanthocyanidins to petal tissue with active hydroxylases resulted in anthocyanidins with increased B-ring hydroxylation relative to the fed leucoanthocyanidin, indicating the presence of 3′-hydroxylating activity (in Petunia and Eustoma grandiflorum Grise.) and 3′,5′-hydroxylating activity (in E. grandiflorum Grise.). Tetcyclacis, a specific inhibitor of cytochrome P450-dependent enzymes, abolished this activity, excluding involvement of unspecific hydroxylases. While some hydroxylation could be a consequence of reverse catalysis by dihydroflavonol 4-reductase (DFR) providing an alternative substrate, hydroxylating activity was still present in fed petals of a DFR deficient petunia line. In vitro conversion rates and kinetic data for dLPG (a stable leucoanthocyanidin substrate) were comparable to those for other flavonoids for nine of ten recombinant flavonoid hydroxylases from various taxa. dLPG was a poor substrate for only the recombinant Fragaria F3′Hs. Thus, the B-ring hydroxylation pattern of anthocyanins can be determined at all precursor levels in the pathway.

Similar content being viewed by others
Abbreviations
- CY:
-
Cyanidin
- DFR:
-
Dihydroflavonol 4-reductase
- dLCy:
-
5-deoxyleucocyanidin
- dLPG:
-
5-deoxyleucopelargonidin
- DP:
-
Delphinidin
- F3´H:
-
Flavonoid 3´-hydroxylase
- F3´5H:
-
Flavonoid 3´5´-hydroxylase
- HBR:
-
Heidi Blue Rim
- LCY:
-
Leucocyanidin
- LPG:
-
Leucopelargonidin
- PG:
-
Pelargonidin
- TET:
-
Tetcyclacis
References
Ausubel F, Bahnsen K, Hanson M, Mitchell AS, Smith HJ (1980) Cell and tissue culture of haploid and diploid petunia ‘Mitchell’. Plant Mol Biol Newsl 1:26–32
Brugliera F, Barri-Rewell G, Holton TA, Mason JG (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19:441–451
Brugliera F, Demelis L, Koes R, Tanaka Y (2010) Genetic sequences having methyltransferases activity and uses thereof. USA Patent US7807877B2, 5 Oct 2010
Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96:12929–12934
Cornu A, Farcy E (1981) Genotype of petunia Mitchell line. Plant Mol Biol Newslett 2:58–109
Davies KM, Marie Bradley J, Schwinn KE, Markham KR, Podivinsky E (1993) Flavonoid biosynthesis in flower petals of five lines of lisianthus (Eustoma grandiflorum Grise.). Plant Sci 95:67–77
Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM (2003) Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131:259–268
De Vlaming P (1981) Genotype determination of Petunia ‘Mitchell’with respect to some flower colour genes. Plant Mol Biol News 2:106–108
Fischer TC, Halbwirth H, Meisel B, Stich K, Forkmann G (2003) Molecular cloning, substrate specificity of the functionally expressed dihydroflavonol 4-reductases from Malus domestica and Pyrus communis cultivars and the consequences for flavonoid metabolism. Arch Biochem Biophys 412:223–230
Forkmann G, Heller W (1999) Biosynthesis of flavonoids. In: Barton D, Nakanishi K, Meth-Cohn O, Sankawa U (eds) Comprehensive natural products chemistry, vol 1. Elsevier Science, Amsterdam, pp 713–748
Forkmann G, Martens S (2001) Metabolic engineering and applications of flavonoids. Curr Opin Biotech 12:155–160
Gosch C, Nagesh KM, Thill J, Miosic S, Plaschil S, Milosevic M, Olbricht K, Ejaz S, Rompel A, Stich K, Halbwirth H (2014) Isolation of dihydroflavonol 4-reductase cDNA clones from Angelonia × angustifolia and heterologous expression as GST fusion protein in Escherichia coli. Plos One, in press
Halbwirth H (2010) The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. Int J Mol Sci 11:595–621
Halbwirth H, Fischer TC, Schlangen K, Rademacher W, Schleifer KJ, Forkmann G, Stich K (2006a) Screening for inhibitors of 2-oxoglutarate-dependent dioxygenases: flavanone 3 [beta]-hydroxylase and flavonol synthase. Plant Sci 171:194–205
Halbwirth H, Kahl S, Jager W, Reznicek G, Forkmann G, Stich K (2006b) Synthesis of (14C)-labeled 5-deoxyflavonoids and their application in the study of dihydroflavonol/leucoanthocyanidin interconversion by dihydroflavonol 4-reductase. Plant Sci 170:587–595
Harborne J (ed) (1976) Functions of flavonoids in plants, vol 1. Chemistry and biochemistry of plant pigments. Academic Press Inc Ltd, London
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Heller W, Britsch L, Forkmann G, Grisebach H (1985a) Leucoanthocyanidins as intermediates in anthocyanidin biosynthesis in flowers of Matthiola incana R. Br Planta 163:191–196
Heller W, Forkmann G, Britsch L, Grisebach H (1985b) Enzymatic reduction of (+)-dihydroflavonols to flavan-3, 4-cis-diols with flower extracts from Matthiola incana and its role in anthocyanin biosynthesis. Planta 165:284–287
Nakajima J-I, Tanaka Y, Yamazaki M, Saito K (2001) Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J Biol Chem 276:25797–25803
Johnson ET, Yi H, Shin B, Oh B-J, Cheong H, Choi G (1999) Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J 19:81–85
Johnson ET, Ryu S, Yi H, Shin B, Cheong H, Choi G (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25:325–333
Kamsteeg J, Brederode JV, Verschuren PM, Nigtevecht GV (1981) Identification, properties and genetic control of p-coumaroylCoenzyme A, 3-hydroxylase isolated from petals of Silene dioica. Z Pflanzenphysiol 102:435–442
Markham KR (1982) Techniques of flavonoid identification. Academic Press, London
Markham KR, Ofman DJ (1993) Lisianthus flavonoid pigments and factors influencing their expression in flower colour. Phytochemistry 34:679–685
Menting JG, Scopes RK, Stevenson TW (1994) Characterization of flavonoid 3′,5′-hydroxylase in microsomal membrane fraction of Petunia hybrida flowers. Plant Physiol 106:633–642
Nielsen KM, Podivinsky E (1997) cDNA cloning and endogenous expression of a flavonoid 3′ 5′-hydroxylase from petals of lisianthus (Eustoma grandiflorum). Plant Sci 129:167–174
Roberts RJ, Vaughan P (1971) Hydroxylation of kaempferol, dihydrokaempferol and naringenin by a phenolase preparation from spinach beet. Phytochemistry 10:2649–2652
Ruhnau B, Forkmann G (1988) Flavan-3, 4-diols in anthocyanin biosynthesis, enzymatic formation with flower extracts from Callistephus chinensis. Phytochemistry 27:1035–1039
Sandermann H, Strominger JL (1972) Purification and properties of C55-isoprenoid alcohol phosphokinase from Staphylococcus aureus. J Biol Chem 247:5123–5131
Schill L, Grisebach H (1973) Properties of a phenolase preparation from cell suspension cultures of parsley. H-S Z Physiol Chem 354:1555–1562
Schlangen K, Miosic S, Topuz F, Muster G, Marosits T, Seitz C, Halbwirth H (2009) Chalcone 3-hydroxylation is not a general property of flavonoid 3′-hydroxylase. Plant Sci 177:97–102
Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381:749–753
Schwinn KE, Markham KR, Giveno NK (1993) Floral flavonoids and the potential for pelargonidin biosynthesis in commercial chrysanthemum cultivars. Phytochemistry 35:145–150
Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G (2006) Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′, 5′-hydroxylase in the Asteraceae family. Plant Mol Biol 61:365–381
Stafford HA, Lester HL, Porter LJ (1985) Chemical and enzymatic synthesis of monomeric procyanidins (leucocyanidins or 3′, 4′, 5, 7-tetrahydroxyflavan-3, 4-diols) from (2R,3R)-dihydroquercetin. Phytochemistry 24:333–338
Stich K, Ebermann R, Forkmann G (1988) Einfluss Cytochrom P-450 spezifischer Inhibitoren auf die Aktivität von Flavonoid 3′-Hydroxylase und Flavonsynthase II bei verschiedenen Pflanzen. Phyton (Austria) 28:237–247
Stotz G, Spribille R, Forkmann G (1984) Flavonoid biosynthesis in flowers of Verbena hybrida. J Plant Physiol 116:173–183
Thill J, Regos I, Farag MA, Ahmad AF, Kusek J, Castro A, Schlangen K, Carbonero CH, Gadjev IZ, Smith LMJ, Halbwirth H, Treutter D, Stich K (2012) Polyphenol metabolism provides a screening tool for beneficial effects of Onobrychis viciifolia (sainfoin). Phytochemistry 82:67–80
Thill J, Miosic S, Gotame TP, Mikulic-Petkovsek M, Gosch C, Veberic R, Preuss A, Schwab W, Stampar F, Stich K (2013) Differential expression of flavonoid 3′-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria × ananassa and Fragaria vesca. Plant Physiol Biochem 72:72–78
Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107:142–149
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Winkel-Shirley B (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107
Acknowledgments
This work was supported by the Austrian Science Fund FWF [Project P24331-B16]. We thank C. Seitz from the former Chair of Floriculture Crops and Horticultural Plant Breeding, Technical University of Munich, Freising for providing some of the F3′H cDNA clones, and W. Rademacher, BASF, for the gift of tecyclacis. We thank C. Martin for the suggestion of including a DFR mutant in the feeding experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special topic: Anthocyanins. Guest editor: Stefan Martens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schwinn, K., Miosic, S., Davies, K. et al. The B-ring hydroxylation pattern of anthocyanins can be determined through activity of the flavonoid 3′-hydroxylase on leucoanthocyanidins. Planta 240, 1003–1010 (2014). https://doi.org/10.1007/s00425-014-2166-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2166-3