Abstract
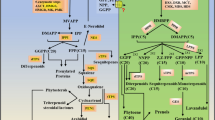
Grapes berries produce and accumulate many reactive secondary metabolites, and encounter a wide range of pathogen- and human-derived xenobiotic compounds. The enzymatic glucosylation of these metabolites changes their reactivity, stability and subcellular location. Two ESTs with more than 90% nucleotide sequence identity to three full-length glucosyltransferases are expressed in several grape tissues. The full-length clones have more than 60% amino acid sequence similarity to previously characterized flavonoid 7-O-glucosyltransferases, catechin O-glucosyltransferases and anthocyanin 5-O-glucosyltransferases. In vitro, these enzymes glucosylate flavonols and the xenobiotic 2,4,5-trichlorophenol (TCP). Kinetic analysis indicates that TCP is the preferred substrate for these enzymes, while expression analysis reveals variable transcription of these genes in grape leaves, flowers and berry tissues. The in vivo role of these Vitis labrusca glucosyltransferases is discussed.




Similar content being viewed by others
Abbreviations
- AF:
-
After flowering
- DFCI:
-
Dana Farber Cancer Institute
- GTs:
-
Glucosyltransferases
- PMSF:
-
Phenylmethyl sulfonyl fluoride
- PSPG:
-
Plant secondary product glucosyltransferases
- PVP-10 or PVPP:
-
Polyvinylpyrrolidone
- Q3G:
-
Quercetin 3-O-glucoside
- TCP:
-
Trichlorophenol
- UDPG:
-
Uridine 5′-diphosphoglucose
- VLOGT:
-
Vitis labrusca O-glucosyltransferase
- VLRSGT:
-
Vitis labrusca resveratrol glucosyltransferase
References
Ali A, Strommer J (2003) A simple extraction and chromatographic system for the simultaneous analysis of anthocyanins and stilbenes of Vitis species. J Agric Food Chem 51:7246–7251
Amico V, Chillemi R, Mangiafico S, Spatafora C, Tringali C (2008) Polyphenol-enriched fractions from Sicilian grape pomace: HPLC-DAD analysis and antioxidant activity. Bioresource Technol 99:5960–5966
Bowles D (2002) A multigene family of glycosyltransferases in a model plant, Arabidopsis thaliana. Biochem Soc Trans 30:301–306
Brazier-Hicks M, Edwards LA, Edwards R (2007) Selection of plants for roles in phytoremediation: the importance of glucosylation. Plant Biotechnol J 5:627–635
Caputi L, Lim E, Bowles D (2008) Discovery of new biocatalysts for the glucosylation of terpenoid scaffolds. Chem Eur J 14:6656–6662
Cascado CG, Heredia A (1999) Structure and dynamics of reconstituted cuticular waxes of grape berry cuticle (Vitis vinifera L.). J Exp Bot 50:175–182
Castillo-Muñoz N, Gómez-Alonso S, García-Romero E, Hermosín-Gutiérrez I (2007) Flavonol profiles of Vitis vinifera red grapes and their single cultivar wines. J Agric Food Chem 55:992–1002
Castillo-Muñoz N, Gómez-Alonso S, García-Romero E, Gómez V, Velders A, Hermosín-Gutiérrez I (2009) Flavonol 3-O-glycosides series of Vitis vinifera cv. Petit Verdot red wine grapes. J Agr Food Chem 57:209–219
Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7:547–552
Fischer T, Gosch C, Pfeiffer J, Halbwirth H, Halle C, Stich K, Forkmann G (2007) Flavonoid genes of pear (Pyrus communis). Trees 21:521–529
Ford CM, Boss PK, Høj PB (1998) Cloning and characterization of Vitis vinifera UDP-Glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273:9224–9233
Hall D, De Luca V (2007) Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J 49:579–591
Harborne JB (1967) Comparative biochemistry of the flavonoids. Academic Press, London
Hirotani M, Kuroda R, Suzuki H, Yoshikawa T (2000) Cloning and expression of UDP-glucose: flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210:1006–1013
Hou B, Lim E, Higgins G, Bowles D (2004) N-Glucosylation of cytokinins by glucosyltransferases of Arabidopsis thaliana. J Biol Chem 279:47822–47832
Huntley A (2007) Grape flavonoids and menopausal health. Menopause Int 13:165–169
Imanishi S, Hashizume K, Kojima H, Ichihara A, Nakamura K (1998) An mRNA of tobacco cell, which is rapidly inducible by methyl jasmonate in the presence of cycloheximide, codes for a putative glycosyltransferases. Plant Cell Physiol 39:202–211
Imayama T, Yoshihara N, Fukuchi-Mizutani M, Tanaka Y, Ino I, Yabuya T (2004) Isolation and characterization of a cDNA clone of UDP-glucose: anthocyanin 5-O-glucosyltransferase in Iris hollandica. Plant Sci 167:1243–1248
Jánváry L, Hoffmann T, Pfeiffer J, Hausmann L, Töpfer R, Fischer T, Schwab W (2009) A double mutation in the anthocyanin 5-O-glucosyltranferase gene disrupts enzymatic activity in Vitis vinifera L. J Agr Food Chem 57:3512–3518
Jones P, Vogt T (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213:164–174
Jones P, Møller B, Høj P (1999) The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor. J Biol Chem 274:35483–35491
Jones P, Meßner B, Nakajima JI, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278:43910–43918
Jurd L (1962) Spectral properties of flavonoid compounds. In: Geissmann TA (ed) The chemistry of flavonoid compounds. Pergamon Press Inc, Oxford, pp 107–155
Kramer CM, Prata RTN, Willits MG, De Luca V, Steffens JC, Graser G (2003) Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry 64:1069–1076
Langcake P, Pryce RJ (1977) A new class of phytoalexins from grapevine. Experientia 33:151–152
Leifert W, Abeywardena M (2008) Cardioprotective actions of grape polyphenols. Nutr Res 28:729–737
Li Y, Baldauf S, Lim EK, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276:4338–4343
Lim E, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid and other benzoates. J Biol Chem 277:586–592
Lim E, Baldauf S, Li Y, Elias L, Worrall D, Spencer SP, Jackson RG, Taguchi G, Ross J, Bowles DJ (2003) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13:139–145
Lim E, Doucet C, Hou B, Jackson R, Abrams S, Bowles D (2005) Resolution of (+)-abscisic acid using an Arabidopsis glucosyltransferase. Tetrahedron Assymetr 16:143–147
Loutre C, Dixon DP, Brazier M, Slater M, Cole DJ, Edwards R (2003) Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant 3, 4-dichloroaniline. Plant J 34:485–493
Meßner B, Thulke O, Schäffner AR (2003) Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217:138–146
Moore MO, Giannasi DE (1994) Foliar flavonoids of eastern North American Vitis (Vitaceae) north of Mexico. Plant Syst Evol 193:21–36
Muñoz-Espada A, Wood K, Bordelon B, Watkins B (2004) Anthocyanin quantification and radical scavenging capacity of Concord, Norton, and Marechal Foch grapes and wines. J Agr Food Chem 52:6779–6786
Nagashima S, Tomo S, Orihara Y, Yoshikawa T (2004) Cloning and characterization of glucosyltransferase cDNA from Eucalyptus perriniana cultured cells. Plant Biotech 21:343–348
Noguchi A, Saito A, Homma Y, Nakao M, Sasaki N, Nishino T, Takahashi S, Nakayama T (2007) A UDP-glucose: isoflavone 7-O-glucosyltransferase from the roots of soybean (Glycine max) seedlings. J Biol Chem 282:23581–23590
Ogata J, Kanno Y, Itoh Y, Tsugawa H, Suzuki M (2005) Plant biochemistry: anthocyanin biosynthesis in roses. Nature 435:757–758
Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T (2010) Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22:2856–2871
Park HJ, Cha HC (2003) Flavonoids from leaves and exocarps of the grape Kyoho. Korean J Biol Sci 7:327–330
Polásková P, Herszage J, Ebeler S (2008) Wine flavor: chemistry in a glass. Chem Soc Rev 37:2478–2489
Schneider E, von der Heydt H, Esperester A (2008) Evaluation of polyphenol composition in red leaves from different varieties of Vitis vinifera. Plant Med 74:565–572
Taguchi G, Ubukata T, Nobuaki H, Yamamoto H, Okazaki M (2003) Cloning and characterization of a glucosyltransferase that reacts on the 7-hydroxyl group of flavonol and the 3-hydroxyl group of coumarin from tobacco cells. Arch Biochem Biophys 420:95–102
Tian L, Blount JW, Dixon RA (2006) Phenylpropanoid glycosyltransferases from osage orange (Maclura pomifera) fruit. FEBS Lett 580:6915–6920
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenow DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5:380–388
Vogt T, Zimmermann E, Grimm R, Meyer M, Strack D (1997) Are the characteristics of betanidin glucosyltransferases from cell-suspension cultures of Dorotheanthus bellidiformis indicative of their phylogenetic relationship with flavonoid glucosyltransferase? Planta 203:349–361
Vogt T, Grimm R, Strack D (1999) Cloning and expression of a cDNA encoding betanidin 5-O-glucosyltransferase, a betanidin- and flavonoid-specific enzyme with high homology to inducible glucosyltransferases from the Solanaceae. Plant J 19:509–519
Willits MG, Giovanni M, Prata RTN, Kramer CM, De Luca V, Steffens JC, Graser G (2004) Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites. Phytochemistry 65:31–41
Wu X, Prior RL (2005) Systematic identification and characterization of anthocyanins by HPLC–ESI–MS/MS in common foods in the United States: fruits and berries. J Agric Food Chem 53:2589–2599
Yamazaki M, Yamagishi E, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K (2002) Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48:401–411
Acknowledgments
This research was funded by a Natural Sciences and Engineering Research Council of Canada Discovery grant to V.D.L. and a Tier 1 Canada Research Chair in Plant Biotechnology to V.D.L., as well as an NSERC post-graduate fellowship to D.H. We thank G&H Wiley Ltd (St, Catharine’s, Ontario, Canada) for allowing us to collect Concord grapes during the growing season. We thank Tim Jones for performing HPLC-mass spectrometry on selected flavonoid reaction products.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2011_1474_MOESM1_ESM.tif
Figure S1 UPLC chromatogram of grape anthocyanins (1, 2, 3, 4, 9) and flavonoids (5, 6, 7, 8) analyzed at 590 and 360 nm, respectively. The Table insert describes the composition of anthocyanins & flavonoids in mature Vitis labrusca grape mesocarp & exocarp. Mean values from 3 biological replicates ± SD are shown (TIFF 11721 kb)
425_2011_1474_MOESM2_ESM.tif
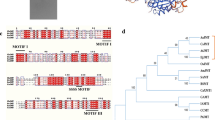
Figure S2 Amino acid sequence alignment of the Vitis labrusca 7-O-glucosyltransferases VLOGT1 (ABQ02256), VLOGT2 (ABQ02257) and VLOGT3 (ABQ02258) with other enzymes which glucosylate the 7-O-position of flavonols. Abbreviations: NtGT2—Nicotiana tabacum coumarin GT (BAB88935), the Lb_catGT- Lycium barbarium catechin GT (BAG80544), Ep_TpGT—Eucalyptus perriniana monoterpene OGT (BAD90934); Ph_5GT—Petunia x hybrida anthocyanin 5-O-glucosyltransferase (BAA89009); Mp_7GT—Maclura pomifera flavonoid 7-O-glucosyltransferase (ABL85474); Pc_7GT—Pyrus communis flavonoid 7-O-glucosyltransferase (AAY27090). The Plant Secondary Product Glucosyltransferase (PSPG) box is underlined. Different colours represent unique amino acids (TIFF 15113 kb)
425_2011_1474_MOESM3_ESM.tif
Figure S3 Neighbor-joining phylogenetic tree of the Vitis labrusca flavonol 7-O-glucosyltransferase gene family and other previously characterized glycosyltransferases. Lb_catGT—Lycium barbarum catechin 4′OGT (BAG80544); Solanum_5gt—Solanum sogarandinum anthocyanin 5OGT (AAK54465); NtGt2—Nicotiana tabacum coumarin GT (BAB88935); Pet_5gt—Petunia x hybrida anthocyanin 5OGT (BAA89009); Pc_7GT—Pyrus communis flavonoid 7OGT (AAY27090); Mp_7gt—Maclura pomifera flavonoid 7OGT (ABL85474); VLOGT3—Vitis labrusca flavonol 7OGT3 (ABQ02258); Vv_TC108954—Vitis vinifera DFCI EST TC108954; Vv_NP9528154—Vitis vinifera genome sequence NP9528154; Vv_LOC100246377—Vitis vinifera genome sequence Locus 100246377; Ep_TpGT1—Eucalyptus perriniana monoterpene GT 1 (BAD90934); Vvx_5GT—Vitis vinifera cv Regent non-functional anthocyanin 5OGT; VV_5GT—Vitis vinifera cv Regent anthocyanin 5OGT; Per_5GT—Perilla frutescens anthocyanin 5OGT (BAA36421); Verb_5GT—Verbena x hybrida anthocyanin 5OGT (BAA36423); Torenia hybrida anthocyanin 5OGT (BAC54093); At_UGT75D1—Arabidopsis thaliana UGT75D1 xenobiotic and flavonol GT; At_UGT75B1—Arabidopsis thaliana UGT75B1 hydroxybenzoic acid OGT; At_UGT75B2—Arabidopsis thaliana UGT75B2 hydroxybenzoic acid OGT; At_UGT75C1_5GT—Arabidopsis thaliana UGT75C1 anthocyanin 5OGT; Vv_TC125631—Vitis vinifera DFCI EST TC125631; Vv_LOC100242998—Vitis vinifera genome sequence Locus 100242998; VLOGT2—Vitis labrusca flavonol 7OGT2 (ABQ02257); Vv_LOC100248120—Vitis vinifera genome sequence Locus 100248120; Vv_ NP9526770- Vitis vinifera genome sequence NP9526770; VLOGT1—Vitis labrusca flavonol 7OGT 1 (ABQ02256); Gent_5GT—Gentiana triflora anthocyanin 5OGT (BAG32255); Iris_5gt—Iris hollandica anthocyanin 5OGT (BAD06874); At_UGT74F2—Arabidopsis thaliana SAGT1 and 4-aminobenzoate OGT; At_UGT74F1—Arabidopsis thaliana UGT74F1 anthranilate OGT; Nt_SAgt—Nicotiana tabacum salicylic acid OGT (AAF61647); Stev_UGT74G1—Stevia rebaudiana UGT74G1 steviol OGT (AAR06920); At_UGT74B1—Arabidopsis thaliana UGT74B1 thiohydroximate SGT; Bn_thiogt—Brassica napus thiohydroximate SGT (AAL09350); At_JAgt—Arabidopsis thaliana UGT74D1 jasmonic acid OGT (NP_180734); Crocus_safgt—Crocus sativus OGT2 (AAP94878); Zea_IAAgt—Zea mays indole-3-acetate OGT (AAA59054); At_UGT84A3—Arabidopsis thaliana UGT84A3 sinapate 1 OGT (NP_193284); At_UGT84A4—Arabidopsis thaliana UGT84A4 sinapate 1 OGT (NP_193285); At_UGT84A2- Arabidopsis thaliana UGT84A2 sinapate 1 OGT (NP_188793); Bn_Sgt1—Brassica napus sinapate glucosyltransferase (AF287143_1); At_UGT84A1—Arabidopsis thaliana sinapate 1 glucosyltransferase (NP_193283); VLRSGT—Vitis labrusca resveratrol/hydroxycinnamic acid OGT (ABH03018); Fra_SAgt2—Fragaria x ananassa cinnamate OGT (AAU09443); Citrus_Limgt—Citrus unshiu liminoid OGT (BAA93039); Mt_UGT84F1—Medicago truncatula isoflavonoid OGT (ABI94023); At_UGT84B1—Arabidopsis thaliana abscisic acid/indole-3-acetic acid OGT/quercetin 7OGT (NP_179907); At_UGT84B2—Arabidopsis thaliana abscisic acid OGT (NP_179906); Dian_chalconegt—Dianthus caryophyllus chalcone 2’OGT (BAD52007); Stev_ugt76G1—Stevia rebaudiana UGT76G1 (AAR06912); At_UGT76E11—Arabidopsis thaliana UGT76E11 quercetin 3OGT and 7OGT (NP_190251); Mt_UGT85H2—Medicago truncatula isoflavonoid OGT (ABI94024); At_UGT85A1_ZeaGT—Arabidopsis thaliana UGT85A1 zeatin OGT (NP_173656); Stev_ugt85C2—Stevia rebaudiana UGT85C2 (AAR06916); Sorghum_UGT85B1—Sorghum bicolor p-hydroxymandelonitrile OGT (Q9SBL1); OryzaIsoFL7GT—Oryza sativa isoflavonoid 7OGT (NP_001059671); Phas_HRA25—Phaseolus vulgaris OGT HRA25 (AF303396_1); Vv_3gt—Vitis vinifera flavonoid 3OGT (AAB81683); VL_3gt—Vitis labrusca flavonoid 3OGT (ABR24135); FraGT1_3GT—Fragaria x ananassa flavonoid 3OGT (AAU09442); At_UGT78D2—Arabidopsis thaliana UGT 78D2 flavonoid 3OGT (NP_197207); At_UGT78D3—Arabidopsis thaliana UGT78D3 flavonol 3-O-arabinosyltransferase (NP_197205); At_UGT78D1—Arabidopsis thaliana UGT 78D1 UDP-rhamnose flavonol 3-O-rhamnosyltransferase (NP_564357); Per_3gt—Perilla frutescens flavonoid 3OGT (BAA19659); Gent _3gt—Gentiana triflora anthocyanidin 3OGT (Q96493); Pet_3gt—Petunia x hybrida anthocyanidin 3OGT (BAA89008); Ipomoea—Ipomoea purpurea flavonoid 3OGT (AAB86473); Mt_UGT78G1—Medicago truncatula isoflavonoid OGT (ABI94025); Dian_3gt—Dianthus caryophyllus flavonol 3OGT (BAD52005); Iris_3gt—Iris hollandica anthocyanidin 3OGT (BAD83701); Hv_3gt—Hordeum vulgare anthocyanidin 3OGT (P14726); Zea_3gt—Zea mays anthocyanidin 3OGT (P16165); Cr_curcgt2—Catharanthus roseus curcumin OGT2 (BAD29722); Gent_3′gt—Gentiana triflora anthocyanin 3’OGT (Q8H0F2); Scut_7gt—Scutellaria baicalensis flavonoid 7OGT (BAA83484); Doro_bet5gt—Dorotheanthus bellidiformis betanidin 5OGT (CAB56231); Rhodiola_salidrosideGT—Rhodiola sachalinensis salidroside OGT (AAS55083); At_UGT73B1—Arabidopsis thaliana UGT73B1 flavonoid 7OGT (NP_567955); Gly_isoflv_gt—Glycyrrhiza echinata isoflavonoid OGT (BAC78438); Vigna_ABAgt—Vigna angularis abscisic acid OGT (BAB83692); Mt_UGT73P1—Medicago truncatula isoflavonoid OGT (ABI94026); Sol_saponin_gt—Solanum aculeatissimum saponin OGT (BAD89042); Sol_sgat1—Solanum tuberosum solanidine galactosyltransferase (AAB48444); At_UGT73C5—Arabidopsis thaliana UGT73C5 zeatin OGT, flavonol 4′ and 7OGT (NP_181218); AtUGT73C6—Arabidopsis thaliana UGT73C6 flavonol 3 and 7OGT (NP_181217); At_UGT73C1—Arabidopsis thaliana UGT73C1 zeatin OGT (NP_181213); Nt_gt4—Nicotiana tabacum flavonoid OGT (BAD93688); Mt_UGT73C8—Medicago truncatula isoflavonoid OGT (ABI94020); At_UGT89B1—Arabidopsis thaliana UGT89B1 flavonol 3,4′ and 7OGT (NP_177529); At_89C1—Arabidopsis thaliana UGT89C1 7-O-rhamnosyltransferase to 7-O-glucose (NP_563756); Mira_DOPAgt—Mirabilis jalapa DOPA 5OGT (BAD91803); Ipo2′′gt—Ipomoea purpurea anthocyanin 2′′OGT (BAD95882); Nt_gt1—Nicotiana tabacum OGT1 (BAB60720); Nt_gt1b—Nicotiana tabacum OGT1b (BAB60721); Nt_gt3—Nicotiana tabacum OGT3 (BAB88934); Cr_curcgt1—Catharanthus roseus curcumin OGT1 (BAD29721); Ipomoea_IAAgt—Ipomoea purpurea indole-3-acetate OGT (BAF75917); Doro_bet6gt—Dorotheanthus bellidiformis betanidin 6OGT (AAL57240); Mt_UGT71G1—Medicago truncatula triterpenoid/flavonoid OGT (2ACW_A); At_UGT71C1—Arabidopsis thaliana UGT71C1 flavonol 3′ and 7OGT (NP_180536); OryzaFl3GT—Oryza sativa flavonol 3OGT (BAC83989); Mt_UGT88E1—Medicago truncatula isoflavonoid OGT (ABI94021); Mt_UGT88E2—Medicago truncatula isoflavonoid OGT (ABI94022); GM_Isoflv7GT—Glycine max isoflavonoid 7OGT (BAF64416); Rose_5gt—Rosa x hybrida anthocyanidin 5,3OGT (Q4R1I9); Antirr_chalcGT—Antirrhinum majus chalcone OGT (BAE48239); Sc_5OGT—Sinningia cardinalis 3-deoxy anthocyanin 5OGT (BAJ11653); Oryza7GT—Oryza sativa flavonoid 7OGT (NP_001044173); Os_RF5—Oryza sativa flavonoid OGT5 (NP_001044170); Bn_UGT2—Brassica napus xenobiotic GT (CAM31954); Mt_epicat_GT—Medicago truncatula epicatechin OGT (ACC38470); Os_RUGT10—Oryza sativa OGT (NP_001174726); Gly_Zeatingt—Glycine max zeatin OGT (AAM09514); Phas_ZOG1—Phaseolus lunatus zeatin OGT (Q9ZSK5); Zea_cisZOGT—Zea mays cis-zeatin OGT (NP_001105017) (TIFF 9949 kb)
Rights and permissions
About this article
Cite this article
Hall, D., Kim, K.H. & De Luca, V. Molecular cloning and biochemical characterization of three Concord grape (Vitis labrusca) flavonol 7-O-glucosyltransferases. Planta 234, 1201–1214 (2011). https://doi.org/10.1007/s00425-011-1474-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1474-0