Abstract
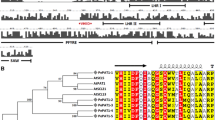
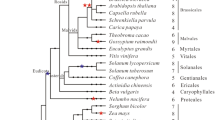
We have cloned and characterized the phytochrome C (PHYC) gene from Stellaria longipes. The PHYC gene is composed of a 110-bp 5′-untranslated leader sequence, a 3,342-bp coding region, and a 351-bp 3′-untranslated sequence. The Stellaria PHYC contains three long introns within the coding region at conserved locations as in most angiosperm PHY genes. DNA blot analysis indicates that the Stellaria genome contains a single copy of PHYC. Stellaria PHYC shares 60%, 58%, and 57% deduced amino acid identities with rice, Sorghum, and Arabidopsis PHYC, respectively. Phylogenetic analysis indicates that Stellaria PHYC is located in the dicot branch, but is divergent from Arabidopsis PHYC. The Stellaria PHYC is constitutively expressed in different plant organs, though the level of PHYC gene transcript in roots is slightly higher than in flowers, leaves, and stems. When 2-week old seedlings grown in the dark were exposed to constant white light, PHYC mRNA quickly accumulates within 1–12 h. When plants grown in darkness for 7 days were exposed to different red/far-red light (R/FR) ratios, the levels of PHYC mRNA at R/FR=0.7 are much lower than under R/FR=3.5. The levels of PHYC mRNA under short-day (SD) photoperiod are higher than under long-day (LD) photoperiod. Plants under SD conditions do not elongate, and are only about 1.7 cm tall at 19 days. In contrast, plants under LD conditions elongate with an average height of 21.2 cm at 19 days. The plants do not flower under SD conditions, but do so at 18–19 days under LD conditions. These results indicate that under SD conditions the high level of PHYC mRNA may inhibit stem elongation and flower initiation. In contrast, under LD conditions the high level of PHYC mRNA may promote stem elongation and flowering.








Similar content being viewed by others
Abbreviations
- LDW :
-
Long-day warm
- PHY :
-
Phytochrome gene
- R/FR :
-
Red/far-red light ratio
- SDC :
-
Short-day cold
References
Alokam S, Chinnappa CC, Reid DM (2002) Red/far-red light mediated stem elongation and anthocyanin accumulation in Stellaria longipes: differential response of alpine and prairie ecotypes. Can J Bot 80:72–81
Altschul F, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aukerman M, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino R, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9:1317–1326
Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH (2000) Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44:27–42
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Casal JJ, Sanchez RA, Yanovsky MJ (1997) The function of phytochrome A. Plant Cell Environ 20:813–819
Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD (1993) Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5:565–575
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25:413–427
Clapham DH, Kolukisaoglu Ü, Larsson CT, Qamaruddin M, Ekberg I, Wiegmann-Eirund C, Schneider-Poetsch HAW, von Arnold S (1999) Phytochrome types in Picea and Pinus. Expression patterns of PHYA-related types. Plant Mol Biol 40:669–678
Cowl JS, Hartley N, Xie DX, Whitelam GC, Murphy GP, Harberd NP (1994) The PHYC gene of Arabidopsis, absence of the third intron found in PHYA and PHYB. Plant Physiol 106:813–814
Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10:1479–1487
Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119:909–915
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull 19:11–15
Eichenberg K, Baurle I, Paulo N, Sharrock RA, Rudiger W, Schafer E (2000) Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett 470:107–112
Emery RJN, Reid DM, Chinnappa CC (1994) Phenotypic plasticity of stem elongation in two ecotypes of Stellaria longipes: the role of ethylene and response to wind. Plant Cell Environ 17:691–700
Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003a) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15:1981–1989
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003b) Phytochrome B, D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131:1340–1346
Hall A, Kozma-Bognar L, Toth R, Nagy F, Millar AJ (2001) Conditional circadian regulation of phytochrome A gene expression. Plant Physiol 127:1808–1818
Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131:1913–1920
Halliday KJ, Thomas B, Whitelam GC (1997) Expression of heterologous phytochromes A, B, or C in transgenic tobacco plants alters vegetative development and flowering time. Plant J 12:1079–1090
Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33:875–885
Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schafer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128:194–200
Hauser BA, Pratt LH, Cordonnier-Pratt MM (1997) Absolute quantification of five phytochrome transcripts in seedlings and mature plants of tomato (Solanum lycopersicum L.). Planta 201:379–387
Hauser BA, Cordonnier-Pratt MM, Pratt LH (1998) Temporal and photoregulated expression of five tomato phytochrome genes. Plant J 14:431–439
Heyer A, Gatz C (1992a) Isolation and characterization of a cDNA-clone coding for potato type A phytochrome. Plant Mol Biol 18:535–544
Heyer A, Gatz C (1992b) Isolation and characterization of a cDNA-clone coding for potato type B phytochrome. Plant Mol Biol 20:589–600
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Kathiresan A, Nagarathna KC, Moloney MM, Reid DM, Chinnappa CC (1998) Differential regulation of 1-aminocyclopropane-1-carboxylate synthase gene family and its role in phenotypic plasticity in Stellaria longipes. Plant Mol Biol 36:265–274
Li WZ, Chinnappa CC (2003) The phytochrome gene family in the Stellaria longipes complex. Int J Plant Sci 164:657–673
Lumsden PJ, Millar AJ (1998) Biological rhythms and photoperiodism in plants. BIOS Scientific, Oxford
Macdonald SE, Chinnappa CC, Reid DM (1984) Studies on Stellaria longipes complex: phenotypic plasticity. I. Response of stem elongation to temperature and photoperiod. Can J Bot 62:414–419
Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13:1141–1150
Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20:666–671
Mathews S, Lavin M, Sharrock RA (1995) Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann Mo Bot Gard 82:296–321
Monte E, Alonso JM, Ecker JR, ZhangY, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling Pathways. Plant Cell 15:1962–1980
Perriere G, Gouy M (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Pratt LH, Cordonnier-Pratt MM, Kelmenson PM, Lazarova GI, Kubota T, Alba RM (1997) The phytochrome gene family in tomato (Solanum lycopersicum L.). Plant Cell Environ 20:672–677
Qin M, Kuhn R, Quail PH (1997) Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J 12:1163–1172
Quail PH (1997a) An emerging molecular map of the phytochromes. Plant Cell Environ 20:657–665
Quail PH (1997b) The phytochromes: a biochemical mechanism of signaling in site? BioEssays 19:571–579
Quail PH (2002) Phytochrome photosensory signaling networks. Nat Rev Mol Cell Biol 3:85–93
Schmidt M, Schneider-Poetsch HA (2002) The evolution of gymnosperms redrawn by phytochrome genes: the Gnetatae appear at the base of gymnosperms. J Mol Evol 54:715–724
Schneider-Poetsch HAW, Kolukisaoglu Ü, Clapham DH, Hughes J, Lamparter T (1998) Non-angiosperm phytochromes and the evolution of vascular plants. Physiol Plant 102:612–622
Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20:840–844
Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616
Weinig C (2002) Phytochrome photoreceptors mediate plasticity to light quality in flowers of the Brassicaceae. Am J Bot 89:230–235
Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20:752–758
Acknowledgement
This work is supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada (A-7222).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, WZ., Chinnappa, C.C. Isolation and characterization of PHYC gene from Stellaria longipes: differential expression regulated by different red/far-red light ratios and photoperiods. Planta 220, 318–330 (2004). https://doi.org/10.1007/s00425-004-1337-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1337-z