Abstract
Objective
Neurocognitive disabilities in Duchenne muscular dystrophy (DMD) children beginning in early childhood and distal DMD gene deletions involving disruption of Dp140 isoform are more likely to manifest significant neurocognitive impairments. MRI data analysis techniques like brain-network metrics can provide information on microstructural integrity and underlying pathophysiology.
Methods
A prospective study on 95 participants [DMD = 57, and healthy controls (HC) = 38]. The muscular dystrophy functional rating scale (MDFRS) scores, neuropsychology batteries, and multiplex ligand-dependent probe amplification (MLPA) testing were used for clinical assessment, IQ estimation, and genotypic classification. Diffusion MRI and network-based statistics were used to analyze structural connectomes at various levels and correlate with clinical markers.
Results
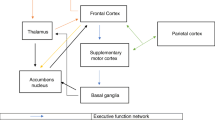
Motor and executive sub-networks were extracted and analyzed. Out of 57 DMD children, 23 belong to Dp140 + and 34 to Dp140- subgroup. Motor disabilities are pronounced in Dp140- subgroup as reflected by lower MDFRS scores. IQ parameters are significantly low in all-DMD cases; however, the Dp140- has specifically lowest scores. Significant differences were observed in global efficiency, transitivity, and characteristic path length between HC and DMD. Subgroup analysis demonstrates that the significance is mainly driven by participants with Dp140- than Dp140 + isoform. Finally, a random forest classifier model illustrated an accuracy of 79% between HC and DMD and 90% between DMD- subgroups.
Conclusions
Current findings demonstrate structural network-based characterization of abnormalities in DMD, especially prominent in Dp140-. Our observations suggest that participants with Dp140 + have relatively intact connectivity while Dp140- show widespread connectivity alterations at global, nodal, and edge levels. This study provides valuable insights supporting the genotype–phenotype correlation of brain-behavior involvement in DMD children.






Similar content being viewed by others
Availability of data and materials
Anonymized data not published within this article will be made available to any appropriately qualified investigator, based on a valid request through the corresponding author.
Code availability (software application or custom code)
Not Applicable.
Abbreviations
- DMD:
-
Duchenne muscular dystrophy
- MRI:
-
Magnetic resonance imaging
- DTI:
-
Diffusion tensor imaging
- FA:
-
Fractional anisotropy
- MD:
-
Mean diffusivity
- MLPA:
-
Multiplex ligation-dependent probe amplification
- MDFRS:
-
Muscular dystrophy functional rating scale
- NBS:
-
Network-based statistics
- SC:
-
Structural connectome
References
Miller G, Wessel HB (1993) Diagnosis of dystrophinopathies: review for the clinician. Pediatr Neurol 9:3–9. https://doi.org/10.1016/0887-8994(93)90002-t
Allen DG, Whitehead NP, Froehner SC (2016) Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol Rev 96:253–305. https://doi.org/10.1152/physrev.00007.2015
Filippo TD, Parisi L, Roccella M (2012) Psychological aspects in children affected by duchenne de boulogne muscular dystrophy. Ment Illn 4:e5. https://doi.org/10.4081/mi.2012.e5
Orsini M, Carolina A, de Ferreira AF et al (2018) Cognitive impairment in neuromuscular diseases: a systematic review. Neurol Int 10:7473. https://doi.org/10.4081/ni.2018.7473
Li G, Jiang W, Du Y, Rossbach K (2017) Intelligence profiles of Chinese school-aged boys with high-functioning ASD and ADHD. Neuropsychiatr Dis Treat 13:1541–1549. https://doi.org/10.2147/NDT.S136477
Naidoo M, Anthony K (2020) Dystrophin Dp71 and the neuropathophysiology of duchenne muscular dystrophy. Mol Neurobiol 57:1748–1767. https://doi.org/10.1007/s12035-019-01845-w
Anand A, Tyagi R, Mohanty M et al (2015) Dystrophin induced cognitive impairment: mechanisms, models and therapeutic strategies. Ann Neurosci 22:108–118. https://doi.org/10.5214/ans.0972.7531.221210
Ashburner J, Friston KJ (2000) Voxel-based morphometry—the methods. Neuroimage 11:805–821. https://doi.org/10.1006/nimg.2000.0582
Fu Y, Dong Y, Zhang C et al (2016) Diffusion tensor imaging study in Duchenne muscular dystrophy. Ann Transl Med 4:109. https://doi.org/10.21037/atm.2016.03.19
Doorenweerd N, Straathof CS, Dumas EM et al (2014) Reduced cerebral gray matter and altered white matter in boys with Duchenne muscular dystrophy. Ann Neurol 76:403–411. https://doi.org/10.1002/ana.24222
Doorenweerd N, Mahfouz A, van Putten M et al (2017) Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Sci Rep 7:12575. https://doi.org/10.1038/s41598-017-12981-5
Preethish-Kumar V, Shah A, Kumar M et al (2020) In Vivo evaluation of white matter abnormalities in children with Duchenne muscular dystrophy using DTI. AJNR Am J Neuroradiol 41:1271–1278. https://doi.org/10.3174/ajnr.A6604
Li J, Shi Y, Toga AW (2016) Mapping brain anatomical connectivity using diffusion magnetic resonance imaging: structural connectivity of the human brain. IEEE Signal Process Mag 33:36–51. https://doi.org/10.1109/MSP.2015.2510024
Shah A, Lenka A, Saini J et al (2017) Altered brain wiring in Parkinson’s disease: a structural connectome-based analysis. Brain Connect 7:347–356. https://doi.org/10.1089/brain.2017.0506
Prasad S, Pandey U, Saini J et al (2019) Atrophy of cerebellar peduncles in essential tremor: a machine learning-based volumetric analysis. Eur Radiol 29:7037–7046. https://doi.org/10.1007/s00330-019-06269-7
Lue Y-J, Su C-Y, Yang R-C et al (2006) Development and validation of a muscular dystrophy-specific functional rating scale. Clin Rehabil 20:804–817. https://doi.org/10.1177/0269215506070809
Steffensen BF, Lyager S, Werge B et al (2002) Physical capacity in non-ambulatory people with Duchenne muscular dystrophy or spinal muscular atrophy: a longitudinal study. Dev Med Child Neurol 44:623–632. https://doi.org/10.1017/s0012162201002663
Oldfield RC (1971) The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9:97–113. https://doi.org/10.1016/0028-3932(71)90067-4
Walker LS, Greene JW (1991) The functional disability inventory: measuring a neglected dimension of child health status. J Pediatr Psychol 16:39–58. https://doi.org/10.1093/jpepsy/16.1.39
Jones-Gotman M (1986) Memory for designs: the hippocampal contribution. Neuropsychologia 24:193–203. https://doi.org/10.1016/0028-3932(86)90052-7
Desikan RS, Ségonne F, Fischl B et al (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980. https://doi.org/10.1016/j.neuroimage.2006.01.021
Behrens TEJ, Berg HJ, Jbabdi S et al (2007) Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage 34:144–155. https://doi.org/10.1016/j.neuroimage.2006.09.018
Rubinov M, Sporns O (2010) Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52:1059–1069. https://doi.org/10.1016/j.neuroimage.2009.10.003
Zalesky A, Fornito A, Bullmore ET (2010) Network-based statistic: identifying differences in brain networks. Neuroimage 53:1197–1207. https://doi.org/10.1016/j.neuroimage.2010.06.041
Pike N (2011) Using false discovery rates for multiple comparisons in ecology and evolution: False discovery rates for multiple comparisons. Methods Ecol Evol 2:278–282. https://doi.org/10.1111/j.2041-210X.2010.00061.x
Bostan AC, Strick PL (2018) The basal ganglia and the cerebellum: nodes in an integrated network. Nat Rev Neurosci 19:338–350. https://doi.org/10.1038/s41583-018-0002-7
Power JD, Cohen AL, Nelson SM et al (2011) Functional network organization of the human brain. Neuron 72:665–678. https://doi.org/10.1016/j.neuron.2011.09.006
Chen Q, Meng Z, Liu X et al (2018) Decision variants for the automatic determination of optimal feature subset in RF-RFE. Genes 9:301. https://doi.org/10.3390/genes9060301
Breiman L (2001) No title found. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Taylor PJ, Betts GA, Maroulis S et al (2010) Dystrophin gene mutation location and the risk of cognitive impairment in Duchenne muscular dystrophy. PLoS ONE 5:e8803. https://doi.org/10.1371/journal.pone.0008803
Jin H, Tan S, Hermanowski J et al (2007) The dystrotelin, dystrophin and dystrobrevin superfamily: new paralogues and old isoforms. BMC Genomics 8:19. https://doi.org/10.1186/1471-2164-8-19
Lidov HG, Selig S, Kunkel LM (1995) Dp140: a novel 140 kDa CNS transcript from the dystrophin locus. Hum Mol Genet 4:329–335. https://doi.org/10.1093/hmg/4.3.329
D’Angelo MG, Lorusso ML, Civati F et al (2011) Neurocognitive profiles in Duchenne muscular dystrophy and gene mutation site. Pediatr Neurol 45:292–299. https://doi.org/10.1016/j.pediatrneurol.2011.08.003
Angelini C, Pinzan E (2019) Advances in imaging of brain abnormalities in neuromuscular disease. Ther Adv Neurol Disord 12:1756286419845567. https://doi.org/10.1177/1756286419845567
Liu J, Li M, Pan Y et al (2017) Complex brain network analysis and its applications to brain disorders: a survey. Complexity 2017:1–27. https://doi.org/10.1155/2017/8362741
Lv S-Y, Zou Q-H, Cui J-L et al (2011) Decreased gray matter concentration and local synchronization of spontaneous activity in the motor cortex in Duchenne muscular dystrophy. AJNR Am J Neuroradiol 32:2196–2200. https://doi.org/10.3174/ajnr.A2718
Burns DP, Roy A, Lucking EF et al (2017) Sensorimotor control of breathing in the mdx mouse model of Duchenne muscular dystrophy. J Physiol (Lond) 595:6653–6672. https://doi.org/10.1113/JP274792
Caudal D, François V, Lafoux A et al (2020) Characterization of brain dystrophins absence and impact in dystrophin-deficient Dmdmdx rat model. PLoS ONE 15:e0230083. https://doi.org/10.1371/journal.pone.0230083
Anderson JL, Head SI, Rae C, Morley JW (2002) Brain function in Duchenne muscular dystrophy. Brain 125:4–13. https://doi.org/10.1093/brain/awf012
Felisari G, Martinelli Boneschi F, Bardoni A et al (2000) Loss of Dp140 dystrophin isoform and intellectual impairment in Duchenne dystrophy. Neurology 55:559–564. https://doi.org/10.1212/wnl.55.4.559
Blitzblau R, Storer EK, Jacob MH (2008) Dystrophin and utrophin isoforms are expressed in glia, but not neurons, of the avian parasympathetic ciliary ganglion. Brain Res 1218:21–34. https://doi.org/10.1016/j.brainres.2008.04.071
Aranmolate A, Tse N, Colognato H (2017) Myelination is delayed during postnatal brain development in the mdx mouse model of Duchenne muscular dystrophy. BMC Neurosci 18:63. https://doi.org/10.1186/s12868-017-0381-0
Carretta D, Santarelli M, Vanni D et al (2003) Cortical and brainstem neurons containing calcium-binding proteins in a murine model of Duchenne’s muscular dystrophy: selective changes in the sensorimotor cortex. J Comp Neurol 456:48–59. https://doi.org/10.1002/cne.10506
Carretta D, Santarelli M, Sbriccoli A et al (2004) Spatial analysis reveals alterations of parvalbumin- and calbindin-positive local circuit neurons in the cerebral cortex of mutant mdx mice. Brain Res 1016:1–11. https://doi.org/10.1016/j.brainres.2004.04.021
Rae MG, O’Malley D (2016) Cognitive dysfunction in Duchenne muscular dystrophy: a possible role for neuromodulatory immune molecules. J Neurophysiol 116:1304–1315. https://doi.org/10.1152/jn.00248.2016
Minciacchi D, Del Tongo C, Carretta D et al (2010) Alterations of the cortico-cortical network in sensori-motor areas of dystrophin deficient mice. Neuroscience 166:1129–1139. https://doi.org/10.1016/j.neuroscience.2010.01.040
Fritschy J-M, Panzanelli P, Tyagarajan SK (2012) Molecular and functional heterogeneity of GABAergic synapses. Cell Mol Life Sci 69:2485–2499. https://doi.org/10.1007/s00018-012-0926-4
Goodnough CL, Gao Y, Li X et al (2014) Lack of dystrophin results in abnormal cerebral diffusion and perfusion in vivo. Neuroimage 102:809–816. https://doi.org/10.1016/j.neuroimage.2014.08.053
Hwa H-L, Chang Y-Y, Chen C-H et al (2007) Multiplex ligation-dependent probe amplification identification of deletions and duplications of the Duchenne muscular dystrophy gene in Taiwanese subjects. J Formos Med Assoc 106:339–346. https://doi.org/10.1016/S0929-6646(09)60318-1
Vengalil S, Preethish-Kumar V, Polavarapu K et al (2017) Duchenne muscular dystrophy and Becker muscular dystrophy confirmed by multiplex ligation-dependent probe amplification: genotype-phenotype correlation in a large cohort. J Clin Neurol 13:91–97. https://doi.org/10.3988/jcn.2017.13.1.91
Doorenweerd N, Dumas EM, Ghariq E et al (2017) Decreased cerebral perfusion in Duchenne muscular dystrophy patients. Neuromuscul Disord 27:29–37. https://doi.org/10.1016/j.nmd.2016.10.005
Doorenweerd N (2020) Combining genetics, neuropsychology and neuroimaging to improve understanding of brain involvement in Duchenne muscular dystrophy—a narrative review. Neuromuscul Disord 30:437–442. https://doi.org/10.1016/j.nmd.2020.05.001
Acknowledgements
We acknowledge and convey our deepest gratitude to all the participants in this study including the young DMD boys, control subjects and their parents, who cooperated impeccably for MRI without even sedation. We also thank our radiographers who went out of the way to help complete the image acquisition of these children patiently even though busy doing regular clinical imaging services.
Funding
This study was partially supported by the educational grant from the “TVS-NIMHANS BURSARY” (grant support no.: TVSB/003/304/2015/00817).
Author information
Authors and Affiliations
Contributions
All authors have contributed in this manuscript, reviewed and approved before submission. No authors have reported any conflict of interested related to this manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was carried out with consent from the appropriate Institutional ethics committee.
Consent to patriciate
The requirement for informed consent was waived-off due to retrospective nature of the study.
Consent for publication
The authors affirm that research participants provided informed consent for publication of the data.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Preethish-Kumar, V., Shah, A., Polavarapu, K. et al. Disrupted structural connectome and neurocognitive functions in Duchenne muscular dystrophy: classifying and subtyping based on Dp140 dystrophin isoform. J Neurol 269, 2113–2125 (2022). https://doi.org/10.1007/s00415-021-10789-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-021-10789-y