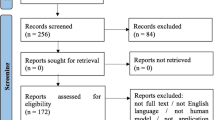
Abstract
Body fluids are one of the most encountered types of evidence in any crime and are commonly used for identifying a person’s identity. In addition to these, they are also useful in ascertaining the nature of crime by determining the ty pe of fluid such as blood, semen, saliva, urine etc. Body fluids collected from crime scenes are mostly found in degraded, trace amounts and/or mixed with other fluids. However, the existing immunological and enzyme-based methods used for differentiating these fluids show limited specificity and sensitivity in such cases. To overcome these challenges, a new method utilizing microRNA expression of the body fluids has been proposed. This method is believed to be non-destructive as well as sensitive in nature and researches have shown promising results for highly degraded samples as well. This systematic review focuses on and explores the use and reliability of miRNAs in body fluid identification. It also summarizes the researches conducted on various aspects of miRNA in terms of body fluid examination in forensic investigations.





Similar content being viewed by others
References
Akutsu T, Watanabe K, Sakurada K (2012) Specificity, sensitivity, and operability of RSID™-Urine for forensic identification of urine: comparison with ELISA for Tamm-Horsfall protein. J Forensic Sci 57(6):1570–1573. https://doi.org/10.1111/j.1556-4029.2012.02174.x
Alshehhi S, Haddrill PR (2020) Evaluating the effect of body fluid mixture on the relative expression ratio of blood-specific RNA markers. Forensic Sci Int 307:110116. https://doi.org/10.1016/j.forsciint.2019.110116
Alshehhi S, Nicola AM, Haddrill PR (2017) Quantification of RNA degradation of blood-specific markers to indicate the age of bloodstains. Forensic Sci International: Genet Supplement Ser 6:e453–e455. https://doi.org/10.1016/j.fsigss.2017.09.175
An JH, Shin KJ, Yang WI, Lee HY (2012) Body fluid identification in forensics. BMB Rep 45(10):545–553. https://doi.org/10.5483/BMBRep.2012.45.10.206
Auvdel ML (1986) Amylase levels in semen and saliva stains. J Forensic Sci 31:426–431. https://doi.org/10.1520/JFS12272J
Bai P, Deng W, Wang L, Long B, Liu K, Liang W, Zhang L (2013) Micro RNA profiling for the detection and differentiation of body fluids in forensic stain analysis. Forensic Sci International: Genet Supplement Ser 4(1):e216–e217. https://doi.org/10.1016/j.fsigss.2013.10.111
Benschop CC, Quaak FC, Boon ME, Sijen T, Kuiper I (2012) Vaginal microbial flora analysis by next generation sequencing and microarrays: can microbes indicate vaginal origin in a forensic context? Int J Legal Med 126:303–310. https://doi.org/10.1007/s00414-011-0660-8
Chong KW, Wong Y, Kiat B, Thong Z, Kiu-Choong Syn C (2015) Development of a RNA profiling assay for biological tissue and body fluid identification. Forensic Sci International: Genet Supplement 5:e196–e198. https://doi.org/10.1016/j.fsigss.2015.09.079
Courts C, Madea B (2011) Specific Micro-RNA signatures for the detection of saliva and blood in forensic body-fluid identification. J Forensic Sci 56(6):1464–1470. https://doi.org/10.1111/j.1556-4029.2011.01894.x
Diamandis EP, Yu H (1975) Non prostatic sources of protein-specific antigen: a steroid hormone dependent phenomenon? Urol Clin North Am 24:275–282. https://doi.org/10.1016/S0094-0143(05)70373-6
Dobay A, Haas C, Fucile G, Downey N, Morrison HG, Kratzer A, Arora N (2019) Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci International: Genet 40:105–113. https://doi.org/10.1016/j.fsigen.2019.02.010
Doi M, Gamo S, Okiura T, Nishimukai H, Asano M (2014) A simple identification method for vaginal secretions using relative quantification of Lactobacillus DNA. Forensic Sci Int Genet 12:93–99. https://doi.org/10.1016/j.fsigen.2014.05.005
Dørum G, Snipen L (2019) Predicting the origin of stains from whole miRNome massively parallel sequencing data. Forensic Sci International: Genet 40:131–139. https://doi.org/10.1016/j.fsigen.2019.02.015
El-Mogy M, Lam B, Haj-Ahmad TA, McGowan S et al (2018) Diversity and signature of small RNA in different bodily fluids using next generation sequencing. BMC Genomics 19(1):1–24. https://doi.org/10.1186/s12864-018-4785-8
Fang C, Zhao J, Li J, Qian J et al (2018) Massively parallel sequencing of microRNA in bloodstains and evaluation of environmental influences on miRNA candidates using realtime polymerase chain reaction. Forensic Sci International: Genet 38:32–38. https://doi.org/10.1016/j.fsigen.2018.10.001
Fleming RI, Harbison SA (2010) The development of an mRNA multiplex RT-PCR assay for the definitive identification of body fluids. Forensic Sci Int Genet 4:244–256. https://doi.org/10.1016/j.fsigen.2009.10.006
Florath I, Butterbach K, Muller H, Bewerunge HM, Brenner H (2014) Cross-sectional and longitudinal changes in DNA methylation state with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet 23:1186–1201. https://doi.org/10.1093/hmg/ddt531
Fujimoto S, Manabe S, Morimoto C, Ozeki M, Hamano Y (2019) Distinct spectrum of microRNA expression in forensically relevant body fluids and probabilistic discriminant approach. Sci Rep 9(1):14332. https://doi.org/10.1038/s41598-019-50796-8
Giampaoli S, Berti A, Valeriani F (2012) Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet 6:559–564. https://doi.org/10.1016/j.fsigen.2012.01.005
Gomes M, Sirker R, Fimmers P, Schneider I (2016) Evaluating the forensic application of 19 target microRNAs as biomarkers in body fluid and tissue identification. Forensic Sci International: Genet 27:41–49. https://doi.org/10.1016/j.fsigen.2016.11.012
Haas C, Hanson E, Kratzer A, Bär W, Ballantyne J (2011) Selection of highly specific and sensitive mRNA biomarkers for the identification of blood. Forensic Sci Int Genet 5:449–458. https://doi.org/10.1016/j.fsigen.2010.09.006
Hanson EK, Lubenow H, Ballantyne J (2009) Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem 387(2):303–314. https://doi.org/10.1016/j.ab.2009.01.037
Hanson EK, Rekab K, Ballantyne J (2013) Binary logistic regression models enable miRNA profiling to provide accurate identification of forensically relevant body fluids and tissues. Forensic Sci International: Genet Supplement Ser 4(1):e127–e128. https://doi.org/10.1016/j.fsigss.2013.10.065
He H, Han N, Ji C, Zhao Y et al (2020) Identification of five types of forensic body fluids based on stepwise discriminant analysis. Forensic Sci International: Genet 48:102337. https://doi.org/10.1016/j.fsigen.2020.102337
He H, Ji A, Zhao Y, Han N et al (2019) A stepwise strategy to distinguish menstrual blood from peripheral blood by Fisher’s discriminant function. Int J Legal Med 134:845–851. https://doi.org/10.1007/s00414-022-02805-1
Hu Z, Lausted C, Yoo H, Yan X et al (2014) Quantitative liver-specific protein fingerprint in blood: a signature for hepatotoxicity. Theranostics 4(2):215
Johnston S, Newman J, Frappier R (2003) Validation study of the Abacus Diagnostics ABAcard® Hematrace® membrane test for the forensic identification of human blood. Can Soc Forensic Sci J 36:173–183. https://doi.org/10.1080/00085030.2003.10757560
Kader F, Ghai M (2020) DNA methylation and application in forensic sciences. Forensic Sci Int 249:255–265
Kind SS (1957) The use of the acid phosphatase test in searching for seminal stains. J Crime Law Criminol Police Sci 47:597–600
Kipps AE, Whitehead PN (1975) The significance of amylase in forensic investigations of body fluids. Forensic Sci 6:137–144. https://doi.org/10.1016/0300-9432(75)90004-7
Kobus HJ, Silenieks E, Scharnberg J (2002) Improving the effectiveness of fluorescence for the detection of Semen stains on fabrics. J Forensic Sci 47(4). https://doi.org/10.1520/JFS15467J
Konrad H, Jurgens L, Hartung B, Poetsch M (2023) More than just blood, saliva, or sperm- setup of a workflow for body fluid identification by DNA methylation analysis. Int J Legal Med 137:1683–1692
Kulstein G, Schacker U, Wiegand P (2018) Old meets new: comparative examination of conventional and innovative RNA-based methods for body fluid identification of laundered seminal fluid stains after modular extraction of DNA and RNA. Forensic Sci International: Genet 36:130–140. https://doi.org/10.1016/j.fsigen.2018.06.017
Layne TR, Green RA, Lewis CA, Nogales F et al (2019) microRNA detection in blood, urine, semen, and Saliva stains after compromising treatments. J Forensic Sci 64(6):1831–1837. https://doi.org/10.1111/1556-4029.14113
Li Z, Bai P, Peng D, Long B et al (2015) Influences of different RT-qPCR methods on forensic body fluid identification by microRNA. Forensic Sci International: Genet Supplement Ser 5:e295–e297. https://doi.org/10.1016/j.fsigss.2015.09.117
Li Z, Bai P, Penga D, Wangb H et al (2017) Screening and confirmation of microRNA markers for distinguishing between menstrual and peripheral blood. Forensic Sci International: Genet 30:24–33. https://doi.org/10.1016/j.fsigen.2017.05.012
Liu Y, He H, Xiao ZX, Ji et al (2020) A systematic analysis of miRNA markers and classification algorithms for forensic body fluid identification. Brief Bioinform 22(4):bbaa324. https://doi.org/10.1093/bib/bbaa324
Lunetta P, Sippel HJ (2009) Positive prostate-specific antigen (PSA) reaction in post mortem rectal swabs: a cautionary note. J Forensic Legal Med 16(7):397–399. https://doi.org/10.1016/j.jflm.2009.04.002
Luo X, Li Z, Peng D, Wang L et al (2015) MicroRNA markers for forensic body fluid identification obtained from miRCURY LNA array. Forensic Sci International: Genet Supplement Ser 5:e630–e632. https://doi.org/10.1016/j.fsigss.2015.10.006
Markland S, Glynn C (2015) Investigating the Simultaneous Extraction of miRNA and DNA from Forensically Relevant Body Fluids. In Conference: Northeastern Association of Forensic Scientists
Mayes C, Houston R, Williams SS, LaRue B, Sheree (2019) The stability and persistence of blood and semen mRNA and miRNA targets for body fluid identification in environmentally challenged and laundered samples. Leg Med 38:45–50. https://doi.org/10.1016/j.legalmed.2019.03.007
Mayes C, Williams SS, Stamm SH (2018) A capillary electrophoresis method for identifying forensically relevant body fluids using miRNAs. Leg Med 30:1–4. https://doi.org/10.1016/j.legalmed.2017.10.013
Misencik A, Laux DL (2007) Validation study of the Seratec HemDirect hemoglobin assay for the forensic identification of human blood. MAFS Newsl 36(2):18–26
Nakanishi H, Kido A, Ohmoris T (2009) A novel method for the identification of saliva by detecting oral streptococci using PCR. Forensic Sci Int 183(1–3):20–23. https://doi.org/10.1016/j.forsciint.2008.10.003
Nunzianda F, Gooch J, Daniel B (2013) Enabling fluorescent biosensors for the forensic identification of body fluids. Analyst 138(24):7279–7288. https://doi.org/10.1039/c3an01372c
O’Leary KR, Glynn CL (2018) Investigating the isolation and amplification of MicroRNAs for forensic body fluid identification. MicroRNA 7(8):187–194. https://doi.org/10.2174/2211536607666180430153821
Omelia EJ, Uchimoto M, Williams G (2013) Quantitative PCR analysis of blood and saliva specific microRNA markers following DNA extraction. Anal Biochem 435(2):120–122. https://doi.org/10.1016/j.ab.2012.12.024
Park JL, Kwon OH, Kim JH (2014) Identification of body fluid-specific DNA methylation markers for use in forensic science. Forensic Sci International: Genet 13:147–153. https://doi.org/10.1016/j.fsigen.2014.07.011
Park JL, Park SM, Kwon OH, Lee H et al (2014) Microarray screening and qRT-PCR evaluation of microRNA markers for forensic body fluid identification. Electrophoresis 35(21–22):3062–3068. https://doi.org/10.1002/elps.201400075
Patel RS, Jakymiw A, Yao B, Pauley BA, Carcamo WC (2012) High resolution of microRNA signatures in human whole saliva. Arch Oral Biol 56(12):1506–1513. https://doi.org/10.1016/j.archoralbio.2011.05.015
Petersen CH, Hjort BB, Tvedebrink T, Kielpinski LJ et al (2013) Body fluid identification of blood, saliva and semen using second generation sequencing of micro-RNA. Forensic Sci International: Genet Supplement Ser 4(1):e204–e205. https://doi.org/10.1016/j.fsigss.2013.10.105
Rhodes C, Campbell A, Lewis C, Seashols SJ (2020) Classification of body fluid source in dried samples using a panel of MicroRNAs (miRNAs). American Academy of Forensic Sciences 72nd Annual Scientific Meeting. American Academy of Forensic Science, Anaheim, California, p 261
Rocchi A, Chiti E, Maiese A, Turillazzi E, Spinett I (2021) MicroRNAs: an update of applications in Forensic Science. Diagnostics 11(1):32. https://doi.org/10.3390/diagnostics11010032
Sauer E, Reinke AK, Courts C (2015) Validation of forensic body fluid identification based on empirically normalized miRNA expression data. Forensic Sci International: Genet Supplement Ser 5:e462–e464. https://doi.org/10.1016/j.fsigss.2015.09.183
Sauer E, Reinke AK, Courts C (2016) Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci International: Genet 22:89–99. https://doi.org/10.1016/j.fsigen.2016.01.018
Simich JP, Morris SL, Klick RL, Diakun R (1999) Validation of the use of a commercially available kit for the identification of prostate specific antigen (PSA) in semen stains. J Forensic Sci 44:1229–1231. https://doi.org/10.1520/JFS14592J
Tian H, Lv M, Li Z, Peng D et al (2017) Semen-specific miRNAs: suitable for the distinction of infertile semen in the body fluid identification? Forensic Sci International: Genet 33:161–167. https://doi.org/10.1016/j.fsigen.2017.12.010
Tobe SS, Watson N, Daéid NN (2007) Evaluation of six presumptive tests for blood, their specificity, sensitivity, and effect on high molecular-weight DNA. J Forensic Sci 52:102–109. https://doi.org/10.1111/j.1556-4029.2006.00324.x
Tong D, Jin Y, Xue T, Ma X et al (2015) Investigation of the application of miR10b and miR135b in the identification of Semen stains. PLoS ONE 10(9):e0137067. https://doi.org/10.1371/journal.pone.0137067
Uchimoto ML, Beasley E, Coult N, Omelia EJ et al (2013) Considering the effect of stem-loop reverse transcription and real-time PCR analysis of blood and saliva specific microRNA markers upon mixed body fluid stains. Forensic Sci International: Genet 7(4):418–421. https://doi.org/10.1016/j.fsigen.2013.04.006
Wang Z, Luo H, Pan X, Liao M, Hou Y (2012) A model for data analysis of microRNA expression in forensic body fluid identification. Forensic Sci International: Genet 6(3):419–423. https://doi.org/10.1016/j.fsigen.2011.08.008
Wang Z, Zhang J, Luo H, Ye Y et al (2013) Screening and confirmation of microRNA markers for forensic body fluid identification. Forensic Sci International: Genet 7(1):116–123. https://doi.org/10.1016/j.fsigen.2012.07.006
Wang Z, Zhang J, Wei W, Zhou D, Luo H (2015) Identification of Saliva using MicroRNA. J Forensic Sci 60(3):702–706. https://doi.org/10.1111/1556-4029.12730
Wang Z, Zhou D, Cao Y, Hu Z et al (2016) Characterization of microRNA expression profiles in blood and saliva using the Ion Personal Genome Machine1 System (Ion PGM System). Forensic Sci International: Genet 20:140–146. https://doi.org/10.1016/j.fsigen.2015.10.008
Weber JA, Baxter DH, Zhang S, Huang DY, Huang K, Jen Lee M, Wang K (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56(11):1733–1741. https://doi.org/10.1373/clinchem.2010.147405
Williams E, Lin MH, Harbison SA, Fleming R (2013) The development of a method for FISH identification of forensically relevant body fluids. Forensic Sci International: Genet Supplement Ser 4(1):107–108. https://doi.org/10.1016/j.fsigss.2013.10.055
Williams G, Uchimoto ML, Coult N, World D, Beasley E (2013) Body fluid mixtures: resolution using forensic microRNA analysis. Forensic Sci International: Genet Supplement Ser 4(1):292–293. https://doi.org/10.1016/j.fsigss.2013.10.149
Williams G, Uchimoto ML, Coult N, World D et al (2013) Characterisation of body fluid specific microRNA markers by capillary electrophoresis. Forensic Sci International: Genet Supplement Ser 4(1):274–275. https://doi.org/10.1016/j.fsigss.2013.10.140
Williams SS, Lewis C, Calloway C, Peace N et al (2016) High-throughput miRNA sequencing and identification of biomarkers for forensically relevant biological fluids. Electrophoresis 37(21):2780–2788. https://doi.org/10.1002/elps.201600258
Yu H, Diamandis EP (1995) Prostate specific antigen in milk of lactating women. Clin Chem 41:54–58. https://doi.org/10.1093/clinchem/41.1.54
Zhao C, Minzhu Z, Zhu Y, Zhang L et al (2021) The persistence and stability of miRNA in bloodstained samples under different environmental conditions. Forensic Sci Int 318:110594. https://doi.org/10.1016/j.forsciint.2020.110594
Zubakov D, Boersma AW, Choi Y, Kuijk PF et al (2010) MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med 124:217–226. https://doi.org/10.1007/s00414-009-0402-3
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
Writing and data analysis: Mohd. Hamza.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Financial interests
The authors have no financial or non-financial interests to disclose.
Clinical trial number
Not applicable.
Competing interests
The authors have no competing interests to declare that are relevant to the article’s content.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hamza, M., Sankhyan, D., Shukla, S. et al. Advances in body fluid identification: MiRNA markers as powerful tool. Int J Legal Med (2024). https://doi.org/10.1007/s00414-024-03202-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00414-024-03202-6