Abstract
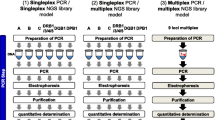
In this study, we assessed to what extent data on the subject of TPPR (transfer, persistence, prevalence, recovery) that are obtained through an older STR typing kit can be used in an activity-level evaluation for a case profiled with a more modern STR kit. Newer kits generally hold more loci and may show higher sensitivity especially when reduced reaction volumes are used, and this could increase the evidential value at the source level. On the other hand, the increased genotyping information may invoke a higher number of contributors in the weight of evidence calculations, which could affect the evidential values as well. An activity scenario well explored in earlier studies [1,2] was redone using volunteers with known DNA profiles. DNA extracts were analyzed with three different approaches, namely using the optimal DNA input for (1) an older and (2) a newer STR typing system, and (3) using a standard, volume-based input combined with replicate PCR analysis with only the newer STR kit. The genotyping results were analyzed for various aspects such as percentage detected alleles and relative peak height contribution for background and the contributors known to be involved in the activity. Next, source-level LRs were calculated and the same trends were observed with regard to inclusionary and exclusionary LRs for persons who had or had not been in direct contact with the sampled areas. We subsequently assessed the impact on the outcome of the activity-level evaluation in an exemplary case by applying the assigned probabilities to a Bayesian network. We infer that data from different STR kits can be combined in the activity-level evaluations.




Similar content being viewed by others
Data availability
Test material is no longer available. Data beyond the details presented in the supplementary data and the Bayesian network in Hugin are sharable upon request and approval of release.
References
Gill P, Hicks T, Butler JM, Connolly E, Gusmão L, Kokshoorn B, Schneider PM (2018) DNA commission of the International society for forensic genetics: assessing the value of forensic biological evidence-Guidelines highlighting the importance of propositions: part i: evaluation of DNA profiling comparisons given (sub-) source propositions. Forensic Sci Int: Genet 36:189–202
Gill P, Hicks T, Butler JM, Connolly E, Gusmão L, Kokshoorn B, Schneider PM (2020) DNA commission of the International society for forensic genetics: assessing the value of forensic biological evidence-Guidelines highlighting the importance of propositions, part II: evaluation of biological traces considering activity level propositions. Forensic Sci Int: Genet 44:102186
Gill P, Benschop C, Buckleton J, Bleka Ø, Taylor D (2021) A review of probabilistic genotyping systems: EuroForMix DNAStatistX and STRmix™. Genes 12(10):1559
van Oorschot RA, Meakin GE, Kokshoorn B, Goray M, Szkuta B (2021) DNA transfer in forensic science: recent progress towards meeting challenges. Genes 12(11):1766
Gosch A, Courts C (2019) On DNA transfer: the lack and difficulty of systematic research and how to do it better. Forensic Sci Int: Genet 40:24–36
Buckingham AK, Harvey ML, van Oorschot RA (2016) The origin of unknown source DNA from touched objects. Forensic Sci Int: Genet 25:26–33
Meakin GE, Kokshoorn B, van Oorschot RA, Szkuta B (2021) Evaluating forensic DNA evidence: connecting the dots. Wiley Interdisciplinary Reviews: Forensic Sci 3(4):e1404
Steensma K, Ansell R, Clarisse L, Connolly E, Kloosterman AD, McKenna LG, Kokshoorn B (2017) An inter-laboratory comparison study on transfer persistence and recovery of DNA from cable ties. Forensic Sci Int: Genet 31:95–104
Goray M, Kokshoorn B, Steensma K, Szkuta B, van Oorschot RA (2020) DNA detection of a temporary and original user of an office space. Forensic Sci Int: Genet 44:102203
Szkuta B, Ansell R, Boiso L, Connolly E, Kloosterman AD, Kokshoorn B, van Oorschot RA (2020) DNA transfer to worn upper garments during different activities and contacts: an inter-laboratory study. Forensic Sci Int: Genet 46:102268
van den Berge M, Ozcanhan G, Zijlstra S, Lindenbergh A, Sijen T (2016) Prevalence of human cell material: DNA and RNA profiling of public and private objects and after activity scenarios. Forensic Sci Int: Genet 21:81–89
van den Berge M, van de Merwe L, Sijen T (2017) DNA transfer and cell type inference to assist activity level reporting: Post-activity background samples as a control in dragging scenario. Forensic Sci Int: Genet Suppl Ser 6:e591–e592
Benschop CC, Hoogenboom J, Hovers P, Slagter M, Kruise D, Parag R, van Marion V (2019) DNAxs/DNAStatistX: Development and validation of a software suite for the data management and probabilistic interpretation of DNA profiles. Forensic Sci Int: Genet 42:81–89
Bhoelai B, Beemster F, Sijen T (2013) Revision of the tape used in a tape-lift protocol for DNA recovery. Forensic Sci Int: Genet Suppl Ser 4(1):e270–e271
Nicklas JA, Buel E (2003) Development of an Alu-based real-time PCR method for quantitation of human DNA in forensic samples. J For Sci 48(5):936–944
Nicklas JA, Buel E (2006) Simultaneous determination of total human and male DNA using a duplex real-time PCR assay. J For Sci 51(5):1005–1015
Lindenbergh A, de Pagter M, Ramdayal G, Visser M, Zubakov D, Kayser M, Sijen T (2012) A multiplex (m) RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int: Genet 6(5):565–577
Duijs F, van de Merwe L, Sijen T, Benschop CC (2018) Low-template methods yield limited extra information for PowerPlex® Fusion 6C profiling. Legal Med 33:62–65
Bleka Ø, Storvik G, Gill P (2016) EuroForMix: an open source software based on a continuous model to evaluate STR DNA profiles from a mixture of contributors with artefacts. Forensic Sci Int: Genet 21:35–44
Bouwman I, Hoogenboom J, Sijen T, Benschop CC (2022) Performing LR calculations when loci are missing between reference and trace DNA profiles. Forensic Sci Int: Reports 5:100268
Tay JW, Murakami JA, Cooper PL, Rye MS (2019) Sensitivity and baseline noise of three new generation forensic autosomal STR kits: PowerPlex® Fusion VeriFilerTM Plus and Investigator® 24plex QS. Forensic Sci Int: Reports 1:100049
Parys-Proszek A, Wróbel M, Marcińska M, Kupiec T (2018) Dual amplification strategy for improved efficiency of forensic DNA analysis using NGM Detect™ NGM™ or Globalfiler™ kits. Forensic Sci Int: Genet 35:46–49
Oldoni F, Podini D (2019) Forensic molecular biomarkers for mixture analysis. Forensic Sci Int: Genet 41:107–119
Taylor D, Biedermann A, Hicks T, Champod C (2018) A template for constructing Bayesian networks in forensic biology cases when considering activity level propositions. Forensic Sci Int: Genet 33:136–146
Benschop CC, Hoogenboom J, Bargeman F, Hovers P, Slagter M, van der Linden J, Sijen T (2020) Multi-laboratory validation of DNAxs including the statistical library DNAStatistX. Forensic Sci Int: Genet 49:102390
Westen AA, Kraaijenbrink T, de Medina EAR, Harteveld J, Willemse P, Zuniga SB, de Knijff P (2014) Comparing six commercial autosomal STR kits in a large Dutch population sample. Forensic Sci Int: Genet 10:55–63
Samie L, Hicks T, Castella V, Taroni F (2016) Stabbing simulations and DNA transfer. Forensic Sci Int: Genet 22:73–80
Steensma K, Ansell R, Clarisse L, Connolly E, Kloosterman AD, McKenna LG, Kokshoorn B (2017) An inter-laboratory comparison study on transfer persistence and recovery of DNA from cable ties. Forensic Sci Int: Genet 31:95–104
Ramos P, Handt O, Taylor D (2020) Investigating the position and level of DNA transfer to undergarments during digital sexual assault. Forensic Sci Int: Genet 47:102316
De Wolff TR, Aarts LHJ, van den Berge M, Boyko T, van Oorschot RAH, Zuidberg M, Kokshoorn B (2021) Prevalence of DNA of regular occupants in vehicles. Forensic Sci Int 320:110713
Acknowledgements
We would like to thank all volunteers for their participation. Margreet van den Berge, Corina Benschop, and Klaas Slooten are thanked for helpful discussions and/or critically reading the manuscript. Jerry Hoogenboom is thanked for programming automized scripts to perform LR calculations.
Author information
Authors and Affiliations
Contributions
Francisca Duijs: investigation, formal analysis, visualization, writing—original draft preparation, and supervision. Erin Meijers: investigation. Bas Kokshoorn: writing—reviewing and editing. Titia Sijen: conceptualization and writing—reviewing and editing. The authors read anda approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Informed consent
Volunteers signed informed consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duijs, F.E., Meijers, E., Kokshoorn, B. et al. Comparison of genotyping and weight of evidence results when applying different genotyping strategies on samples from a DNA transfer experiment. Int J Legal Med 137, 47–56 (2023). https://doi.org/10.1007/s00414-022-02918-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-022-02918-7