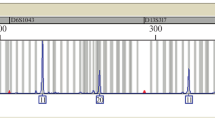
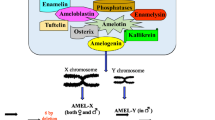
Abstract
Gender identification in forensic DNA typing is an important tool for criminal investigation as well as STR typing. Most methods are based on a size difference of amelogenin X and Y (AMELX and AMELY). There have been some reports that the method by amelogenin (AMEL) incorrectly typed some males as females because the AMELY allele is not detected. AMELY allele dropout is often caused by deletions encompassing AMELY on Yp11.2 and accompanied with Y-STR allele dropout (especially DYS458). However, an unusual deletion was found in our laboratory. The AMELY allele could be recovered by using another commercial kit and alternative AMEL primer sets, and the Y-STR markers resulted in a complete profile. Sequencing results showed that there was an 8 bp deletion in AMELY at the position corresponding to 41–48 bp downstream from the 3′ end of the 6 bp deletion site in AMELX. This is considered a novel mutation at a primer binding region.





Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Sullivan KM, Mannucci A, Kimpton CP, Gill P (1993) A rapid and quantitative DNA sex test: fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques 15:636–641
Santos FR, Pandya A, Tyler-Smith PC (1998) Reliability of DNA based sex tests. Nature Genet 18:103. https://doi.org/10.1038/ng0298-103
Lattanzi W, Di Giacomo MC, Lenato GM, Chimienti G, Voglino G, Resta N, Pepe G, Guanti G (2005) A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum Genet 116:395–401. https://doi.org/10.1007/s00439-004-1238-z
Jobling MA, Lo ICC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, Chang YM, Kraaijenbrink T, Henke J, Guanti G, McKeown B, van Oorschot RAH, Mitchell RJ, de Knijff P, Tyler-Smith C, Parkin EJ (2007) Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet 16:307–316. https://doi.org/10.1093/hmg/ddl465
Chang YM, Perumal R, Keat PY, Yong RYY, Kuehn DLC, Burgoyne L (2007) A distinct Y-STR haplotype for Amelogenin negative males characterized by a large Yp11.2 (DYS458-MSY1-AMEL-Y) deletion. Forensic Sci Int 166:115–120. https://doi.org/10.1016/j.forsciint.2006.04.013
Yong RYY, Gan LSH, Chang YM, Yap PHY (2007) Molecular characterization of a polymorphic 3-Mb deletion at chromosome Yp11.2 containing the AMELY locus in Singapore and Malaysia populations. Hum Genet 122:237–249. https://doi.org/10.1007/s00439-007-0389-0
Kumagai R, Sasaki Y, Tokuta T, Biwasaka H, Aoki Y (2008) DNA analysis of family members with deletion in Yp11.2 region containing amelogenin locus. Leg Med 10:39–42. https://doi.org/10.1016/j.legalmed.2007.05.009
Ou X, Chen W, Chen H, Zhao F, Zheng J, Tong D, Chen Y, Chen A, Sun H (2012) Null alleles of the X and Y chromosomal amelogenin gene in a Chinese population. Int J Legal Med 126:513–518. https://doi.org/10.1007/s00414-011-0594-1
Ma Y, Kuang JZ, Zhang J, Wang GM, Wang YJ, Jin WM, Hou YP (2012) Y chromosome interstitial deletion induced Y-STR allele dropout in AMELY-negative individuals. Int J Legal Med 126:713–724. https://doi.org/10.1007/s00414-012-0720-8
Borovko S, Shyla A, Korban V, Borovko A (2015) Amelogenin test abnormalities revealed in Belarusian population during forensic DNA analysis. Forensic Sci Int Genet 15:98–104. https://doi.org/10.1016/j.fsigen.2014.10.014
Takayama T, Takada N, Suzuki R, Nagaoka S, Watanabe Y, Kumagai R, Aoki Y, Butler JM (2009) Determination of deleted regions from Yp11.2 of an amelogenin negative male. Leg Med 11:S578–S580. https://doi.org/10.1016/j.legalmed.2009.01.049
Cheng JB, Liu Q, Long F, Huang DX, Yi SH (2019) Analysis of the Yp11.2 deletion region of phenotypically normal males with an AMELY-Null allele in the Chinese Han population. Genet Test Mol Biomarkers 23:359–362. https://doi.org/10.1089/gtmb.2018.0231
Krenke BE, Tereba A, Anderson SJ, Buel E, Culhane S, Finis CJ, Tomsey CS, Zachetti JM, Masibay A, Rabbach DR, Amiott EA, Sprecher CJ (2002) Validation of a 16-locus fluorescent multiplex system. J Forensic Sci 47:773–785
Eng B, Ainsworth P, Waye JS (1994) Anomalous migration of PCR products using nondenaturing polyacrylamide gel electrophoresis: the amelogenin sex-typing system. J Forensic Sci 39:1356–1359
Chen W, Wu W, Cheng J, Zhang Y, Chen Y, Sun H (2014) Detection of the deletion on Yp11.2 in a Chinese population. Forensic Sci Int Genet 8:73–79. https://doi.org/10.1016/j.fsigen.2013.07.003
Shadrach B, Commane M, Hren C, Warshawsky I (2004) A rare mutation in the primer binding region of the amelogenin gene can interfere with gender identification. J Mol Diag 6:401–405. https://doi.org/10.1016/S1525-1578(10)60538-7
Maciejewska A, Pawłowski R (2009) A rare mutation in the primer binding region of the Amelogenin X homologue gene. Forensic Sci Int Genet 3:265–267. https://doi.org/10.1016/j.fsigen.2009.01.010
Drobnic K (2006) A new primer set in a SRY gene for sex identification. Int Cong Series 1288:268–270. https://doi.org/10.1016/j.ics.2005.08.020
Morikawa T, Yamamoto Y, Miyaishi S (2011) A new method for sex determination based on detection of SRY, STS and amelogenin gene regions with simultaneous amplification of their homologous sequences by a multiplex PCR. Acta Med Okayama 65: 113–122. http://doi.org/https://doi.org/10.18926/AMO/45270.
Masuyama K, Shojo H, Nakanishi H, Inokuchi S, Adachi N (2017) Sex determination from fragmented and degenerated DNA by amplified product-length polymorphism bidirectional SNP analysis of amelogenin and SRY genes. PLoS ONE 12:e0169348. https://doi.org/10.1371/journal.pone.0169348
Pereira ET, de Almeida JC, Gunha AC, Patton M, Taylor R, Jeffery S (1991) Use of probes for ZFY, SRY, and the Y pseudoautosomal boundary in XX males, XX true hermaphrodites, and an XY female. J Med Genet 28:591–595. https://doi.org/10.1136/jmg.28.9.591
Casas-Vargas A, Galvis J, Blanco J, Rengifo L, Usaquén W, Velasco H (2019) Male patient 46, XX SRY-negative and unambiguous genitalia: a case report. Biomedica 39:622–630. https://doi.org/10.7705/biomedica.4687
Mullaney JM, Mills RE, Pittard WS, Devine SE (2010) Small insertions and deletions (INDELs) in human genomes. Hum Mo Genet 19:R131–R136. https://doi.org/10.1093/hmg/ddq400
Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE (2006) An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 16:1182–1190. http://www.genome.org/cgi/doi/https://doi.org/10.1101/gr.4565806.
Acknowledgements
I would like to thank Dr. John M. Butler, Special Assistant to the Director for Forensic Science, NIST Office of Special Programs, for his valuable comments.
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Ethics approval, consent to participate, consent for publication
Although ethics approval was not obtained, when collecting materials, the content and purpose of the test were fully explained to the material donor, and written informed consent was obtained. In addition, this is a case report as part of forensic casework, and sufficient care has been taken to ensure that no individual or case can be identified.
Conflict of interest
The author declares no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Takayama, T. A novel mutation at the AMEL primer binding region on the Y chromosome in AMELY negative male. Int J Legal Med 136, 519–526 (2022). https://doi.org/10.1007/s00414-022-02781-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-022-02781-6