Abstract
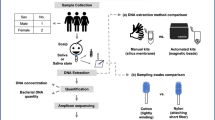
The identification of biological traces provides vital evidence in forensic reconstruction at crime scenes, especially in sexual offences. Compared with traditional presumptive or confirmatory methods, the microbiome-based method has been proven to be of great value in body fluid identification. Mixture of body fluids or tissue is common in sexual assault cases; thus, it is essential to determine the sources of mixed samples. In this study, 60 samples consisting of skin, saliva, and a mixed model of saliva deposited on facial skin were collected from a population living in Guangdong. Through 16s rDNA high-throughput sequencing, we identified the predominant microbes in saliva samples, viz., Haemophilus parainfluenzae T3T1, Neisseria flava, Gemella haemolysans, Prevotella melaninogenica, and Actinomyces odontolyticus; in skin samples, Cutibacterium acnes and Corynebacterium tuberculostearicum were the predominant species. The microbial composition of the same body fluid or tissue is similar in different individuals. However, among different body fluids or tissue, the composition of microflora in saliva is more stable than that on skin. Additionally, the microbial community in the mixed model of saliva deposited on facial skin from the same and different individuals was clearly determined by the constituent fluids or tissue, apart from the differences among the donors. Overall, the microbiome-based method may have good potential as a tool for identifying single and mixed body fluid or tissue.







Similar content being viewed by others
References
An JH, Shin KJ, Yang WI, Lee HY (2012) Body fluid identification in forensics. BMB Rep 45(10):545–553
Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188(1-3):1–17
Sijen T (2015) Molecular approaches for forensic cell type identification: on mRNA, miRNA, DNA methylation and microbial markers. Forensic Sci Int Genet 18:21–32
Oliveira M, Amorim A (2018) Microbial forensics: new breakthroughs and future prospects. Appl Microbiol Biotechnol 102(24):10377–10391
Stahl DA, Davidson SK (2006) Blueprints for partnerships. Nature 443(7114):925–927
Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207–214
Lax S, Hampton-Marcell JT, Gibbons SM, et al (2015) TForensic analysis of the microbiome of phones and shoes. Microbiome 3:21
Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol 8(6):e1002358
Diez Lopez C, Vidaki A, Ralf A, Montiel Gonzalez D, Radjabzadeh D, Kraaij R, Uitterlinden AG, Haas C, Lao O, Kayser M (2019) Novel taxonomy-independent deep learning microbiome approach allows for accurate classification of different forensically relevant human epithelial materials. Forensic Sci Int Genet 41:72–82
Zou KN, Ren LJ, Ping Y, Ma K, Li H, Cao Y, Zhou HG, Wei YL (2016) Identification of vaginal fluid, saliva, and feces using microbial signatures in a Han Chinese population. J Forensic Legal Med 43:126–131
Huang H, Yao T, Wu W, Zhai C, Guan T, Song Y, Sun Y, Xiao C, Liang P, Chen L (2019) Specific microbes of saliva and vaginal fluid of Guangdong Han females based on 16S rDNA high-throughput sequencing. Int J Legal Med 133(3):699–710
Dobay A, Haas C, Fucile G, Downey N, Morrison HG, Kratzer A, Arora N (2019) Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci Int Genet 40:105–113
Tackmann J, Arora N, Schmidt TSB, Rodrigues JFM, von Mering C (2018) Ecologically informed microbial biomarkers and accurate classification of mixed and unmixed samples in an extensive cross-study of human body sites. Microbiome 6(1):192
Hanssen EN, Avershina E, Rudi K, Gill P, Snipen L (2017) Body fluid prediction from microbial patterns for forensic application. Forensic Sci Int Genet 30:10–17
Soares-Vieira JA, Billerbeck AE, Iwamura ES, Zampieri RA, Gattas GJ, Munoz DR, Hallak J, Mendonca BB, Lucon AM (2007) Y-STRs in forensic medicine: DNA analysis in semen samples of azoospermic individuals. J Forensic Sci 52(3):664–670
Kenna J, Smyth M, McKenna L, Dockery C, McDermott SD (2011) The recovery and persistence of salivary DNA on human skin. J Forensic Sci 56(1):170–175
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC (2017) Accuracy of microbial community diversity estimated by closed- and open-reference OTUs. PeerJ 5:e3889
Chung NC, Miasojedow B, Startek M, Gambin A (2019) Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data. BMC Bioinformatics 20(Suppl 15):644–644
Byrd AL, Belkaid Y, Segre JA (2018) The human skin microbiome. Nat Rev Microbiol 16(3):143–155
Oh J, Byrd AL, Deming C, Conlan S, NCS Program, Kong HH, Segre JA (2014) Biogeography and individuality shape function in the human skin metagenome. Nature 514(7520):59–64
Zaura E, Keijser BJ, Huse SM, Crielaard W (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259
Fyhrquist N, Muirhead G, Prast-Nielsen S, Jeanmougin M, Olah P, Skoog T, Jules-Clement G, Feld M, Barrientos-Somarribas M, Sinkko H, van den Bogaard EH, Zeeuwen P, Rikken G, Schalkwijk J, Niehues H, Daubener W, Eller SK, Alexander H, Pennino D, Suomela S, Tessas I, Lybeck E, Baran AM, Darban H, Gangwar RS, Gerstel U, Jahn K, Karisola P, Yan L, Hansmann B, Katayama S, Meller S, Bylesjo M, Hupe P, Levi-Schaffer F, Greco D, Ranki A, Schroder JM, Barker J, Kere J, Tsoka S, Lauerma A, Soumelis V, Nestle FO, Homey B, Andersson B, Alenius H (2019) Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun 10(1):4703
Kennedy B, Peura S, Hammar U, Vicenzi S, Hedman A, Almqvist C, Andolf E, Pershagen G, Dicksved J, Bertilsson S, Fall T (2019) Oral microbiota development in early childhood. Sci Rep 9(1):19025
Older CE, Diesel AB, Lawhon SD, Queiroz CRR, Henker LC, Rodrigues Hoffmann A (2019) The feline cutaneous and oral microbiota are influenced by breed and environment. PLoS One 14(7):e0220463
Phan K, Barash M, Spindler X, Gunn P, Roux C (2020) Retrieving forensic information about the donor through bacterial profiling. Int J Legal Med 134(1):21–29
Ohta J, Noda N, Minegishi S, Sakurada K (2019) Application of DNA repair for Streptococcus salivarius DNA-based identification of saliva from ultraviolet-exposed samples. Forensic Sci Int 306:110077
Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT (2014) Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9(4):e93827
Ling Z, Liu X, Wang Y, Li L, Xiang C (2013) Pyrosequencing analysis of the salivary microbiota of healthy Chinese children and adults. Microb Ecol 65(2):487–495
Willis JR, Gonzalez-Torres P, Pittis AA, Bejarano LA, Cozzuto L, Andreu-Somavilla N, Alloza-Trabado M, Valentin A, Ksiezopolska E, Company C, Onywera H, Montfort M, Hermoso A, Iraola-Guzman S, Saus E, Labeeuw A, Carolis C, Hecht J, Ponomarenko J, Gabaldon T (2018) Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome 6(1):218
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA (2009) Topographical and temporal diversity of the human skin microbiome. Science 324(5931):1190–1192
Grice EA (2014) The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg 33(2):98–103
Acknowledgments
We are grateful to all volunteers who contributed samples for this study.
Funding
This project was supported by the National Natural Science Foundation of China (Grant no. 81501627 and Grant no. 81930055) and Natural Science Foundation of Guangdong Province (Grant no. 2020A1515010938).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The project was approved by the biomedical ethical committee of the Southern Medical University (No. 2019-0011).
Conflict of Interest
The authors declare that they have no conflict of interest.
Informed consent
All study volunteers have signed the informed consent forms before they entered the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1. In this study, a mixture of saliva deposited on facial skin was used to simulate mixed models, and microbial community of mixture was clearly dominated by the constituent fluid or tissue.
2. The microbial composition of skin and saliva were significantly different, and the microbes with significant abundance differences were listed.
3. Microbiome-based method could be useful in identification of skin, saliva, and mixture of them.
Electronic supplementary material
Supplementary Table 1
(DOCX 24 kb).
Rights and permissions
About this article
Cite this article
Yao, T., Han, X., Guan, T. et al. Exploration of the microbiome community for saliva, skin, and a mixture of both from a population living in Guangdong. Int J Legal Med 135, 53–62 (2021). https://doi.org/10.1007/s00414-020-02329-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-020-02329-6