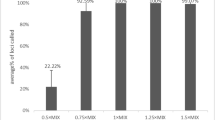
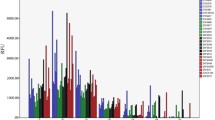
Abstract
The SureID® PathFinder Plus is a new 6-dye, 41-plex Y-STR system that includes the 17 loci from the Yfiler® kit (DYS19, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, and Y-GATA-H4) plus 14 rapidly mutating Y-STR loci (DYS449, DYS481, DYS518, DYS527a/b, DYS533, DYS549, DYS570, DYS576, DYS627, DYF387S1a/b, and DYF404S1), and 10 low-medium mutation loci (DYS388, DYS444, DYS447, DYS460, DYS522, DYS557, DYS593, DYS596, DYS643, and DYS645). The inclusion of the 14 rapidly mutating Y-STR loci improves the discrimination of related individuals. Conversely, the 10 low-medium mutation loci are suitable not only for familial searching but also for providing a higher refinement in the construction of Y chromosome phylogenetic relationships among lineages. The 41-plex Y-STR system is designed for direct amplification of reference samples, such as blood samples on an FTA® Card, gauze, tissue, or cotton substrates as well as hair root or buccal samples on swabs. We performed developmental validation work including accuracy, stability, stutter precision, species specificity, sensitivity, PCR inhibitors, reproducibility, parallel testing of the system, and suitability for use on DNA mixtures. In addition, mutations of the loci were analyzed by 754 DNA-confirmed father–son pairs. The results demonstrate that this kit, developed in-house, is time-efficient, accurate, reliable, and highly informative for forensic database, familial searching, and distinguishing related males.




Similar content being viewed by others
References
Gopinath S, Zhong C, Nguyen V, Ge J, Lagace RE, Short ML, Mulero JJ (2016) Developmental validation of the Yfiler® Plus PCR Amplification Kit: an enhanced Y-STR multiplex for casework and database applications. Forensic Sci Int Genet 24:164–175
Hedman M, Pimenoff V, Lukka M, Sistonen P, Sajantila A (2004) Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages. Forensic Sci Int 142:37–43
Hedman M, Neuvonen AM, Sajantila A, Palo JU (2010) Dissecting the Finnish male uniformity: the value of additional Y-STR loci. Forensic Sci Int Genet 5:199–201
Kayser M, Vermeulen M, Knoblauch H, Schuster H, Krawczak M, Roewer L (2007) Relating two deep-rooted pedigrees from Central Germany by high-resolution Y-STR haplotyping. Forensic Sci Int Genet 1:125–128
Lin H, Ye Q, Tang P, Mo T, Yu X, Tang J (2020) Analyzing genetic polymorphism and mutation of 44 Y-STRs in a Chinese Han population of Southern China. Legal Med 42:101643
Thompson JM, Ewing MM, Frank WE, Pogemiller JJ, Nolde CA, Koehler DJ, Shaffer AM, Rabbach DR, Fulmer PM, Sprecher CJ, Storts DR (2012) Developmental validation of the PowerPlex® Y23 System: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci Int Genet 7:240–250
Westen AA, Kraaijenbrink T, Clarisse L, Grol LJW, Willemse P, Zuniga SB, Robles De Medina EA, Schouten R, van der Gaag KJ, Weiler NEC, Kal AJ, Kayser M, Sijen T, de Knijff P (2014) Analysis of 36 Y-STR marker units including a concordance study among 2085 Dutch males. Forensic Sci Int Genet 14:174–181
Ballantyne KN, Keerl V, Wollstein A, Choi Y, Zuniga SB, Ralf A, Vermeulen M, de Knijff P, Kayser M (2012) A new future of forensic Y-chromosome analysis: rapidly mutating Y-STRs for differentiating male relatives and paternal lineages. Forensic Sci Int Genet 6:208–218
Butler JM, Schoske R (2005) U.S. population data for the multi-copy Y-STR locus DYS464. J Forensic Sci 50:975
Robino C, Ralf A, Pasino S, De Marchi MR, Ballantyne KN, Barbaro A, Bini C, Carnevali E, Casarino L, Di Gaetano C, Fabbri M, Ferri G, Giardina E, Gonzalez A, Matullo G, Nutini AL, Onofri V, Piccinini A, Piglionica M, Ponzano E, Previderè C, Resta N, Scarnicci F, Seidita G, Sorçaburu-Cigliero S et al (2014) Development of an Italian RM Y-STR haplotype database: results of the 2013 GEFI collaborative exercise. Forensic Sci Int Genet 15:56–63
Mo XT, Zhang J, Ma WH, Bai X, Li WS, Zhao XC, Ye J (2019) Developmental validation of the DNATyperTM Y26 PCR amplification kit: an enhanced Y-STR multiplex for familial searching. Forensic Sci Int Genet 38:113–120
Mulero JJ, Chang CW, Calandro LM, Green RL, Li Y, Johnson CL, Hennessy LK (2006) Development and validation of the AmpFlSTR® YfilerTM PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci 51:64–75
Shi MS, Tang JP, Bai RF, Yu XJ, Lv JY, Hu B (2007) Haplotypes of 20 Y-chromosomal STRs in a population sample from southeast China (Chaoshan area). Int J Legal Med 121:455–462
Dai HL, Wang XD, Li YB, Wu J, Zhang J, Zhang HJ, Dong JG, Hou YP (2004) Characterization and haplotype analysis of 10 novel Y-STR loci in Chinese Han population. Forensic Sci Int 145:47–55
Hanson EK, Ballantyne J (2007) Population data for 48 'Non-Core' Y chromosome STR loci. Legal Med 9:221–231
Jarve M, Zhivotovsky LA, Rootsi S, Help H, Rogaev EI, Khusnutdinova EK, Kivisild T, Sanchez JJ (2009) Decreased rate of evolution in Y chromosome STR loci of increased size of the repeat unit. PLoS One 4:e7276
Du W, Feng P, Huang H, Wu W, Zhang L, Guo Y, Liu C, Liu H, Liu C, Chen L (2019) Technical note: developmental validation of a novel 6-dye typing system with 36 Y-STR loci. Int J Legal Med 133:1015–1027
Alghafri R, Goodwin W, Ralf A, Kayser M, Hadi S (2015) A novel multiplex assay for simultaneously analysing 13 rapidly mutating Y-STRs. Forensic Sci Int Genet 17:91–98
Baeta M, Nunez C, Villaescusa P, Ortueta U, Ibarbia N, Herrera RJ, Blazquez-Caeiro JL, Builes JJ, Jimenez-Moreno S, Martinez-Jarreta B, de Pancorbo MM (2018) Assessment of a subset of Slowly Mutating Y-STRs for forensic and evolutionary studies. Forensic Sci Int Genet 34:e7–e12
Ballantyne K, Ralf A, Aboukhalid R, Achakzai N, Anjos T, Ayub Q, Balažic J, Ballantyne J, Ballard D, Berger B, Bobillo C, Bouabdellah M, Burri H, Capal T, Caratti S, Cárdenas J, Cartault F, Carvalho E, Carvalho M, Cheng B, Coble M, Comas D, Corach D, D'Amato M, Davison S et al (2014) Toward male individualization with rapidly mutating Y-chromosomal short tandem repeats. Hum Mutat 35:1021–1032
SWGDAM (2004) Report on the current activities of the Scientific Working Group on DNA Analysis Methods Y-STR Subcommitee. Available at http://www2.fbi.gov/hq/lab/fsc/backissu/july2004/standards/2004_03_standards03.htm. Accessed 10.12.12
SWGDAM (2012) Validation Guidelines for DNA Analysis Methods. Available at http://swgdam.org/SWGDAM_Validation_Guidelines_APPROVED_Dec_2012.pdf. Accessed 04.05.15
SWGDAM (2014) Interpretation Guidelines for Y-Chromosome STR Typing. Available at http://swgdam.org/SWGDAM_YSTR_Guidelines_APPROVED_01092014_v_02112014_FINAL.pdf. Accessed 04.05.15
Nei M, Tajima F (1981) DNA polymorphism detectable by restriction endonucleases. Genetics 97:145–163
Hedman J, Nordgaard A, Rasmusson B, Ansell R, Radstrom P (2009) Improved forensic DNA analysis through the use of alternative DNA polymerases and statistical modeling of DNA profiles. Biotechniques 47:951–958
Opel KL, Chung D, McCord BR (2010) A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci 55:25–33
Tsai YL, Olson BH (1992) Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol 58:2292–2295
Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K (1994) Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci 39:362
Shutler GG, Gagnon P, Verret G, Kalyn H, Korkosh S, Johnston E, Halverson J (1999) Removal of a PCR inhibitor and resolution of DNA STR types in mixed human-canine stains from a five year old case. J Forensic Sci 44:623–626
Schlotterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20:211–215
Viguera E, Canceill D, Ehrlich SD (2001) In vitro replication slippage by DNA polymerases from thermophilic organisms. J Mol Biol 312:323–333
Walsh PS, Fildes NJ, Reynolds R (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res 24:2807–2812
Acknowledgments
We thank all of the participants in this study. The authors would like to gratefully acknowledge the following laboratories for beta-testing of the SureID® PathFinder Plus Kit: Medical Research Center of Shaoxing University, Forensic Center of Shaoxing University, Human DNA Diagnostics Laboratory of Ningbo Health Gene Technologies Co. Ltd., and Forensic DNA laboratory of Shaoxing Municipal Public Security Bureau.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent was obtained from all participating subjects, and this work was approved by the Ethics Committee of the Medical College, Shaoxing University.
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 45683 kb).
Rights and permissions
About this article
Cite this article
Fan, G., Pan, L., Tang, P. et al. Technical note: developmental validation of a novel 41-plex Y-STR system for the direct amplification of reference samples. Int J Legal Med 135, 409–419 (2021). https://doi.org/10.1007/s00414-020-02326-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-020-02326-9