Abstract
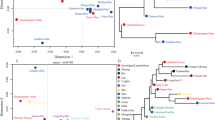
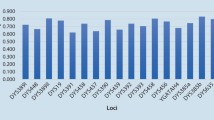
Haplotype diversity for 23 Y chromosomal short tandem repeat (Y-STR) loci included in the PowerPlex® Y23 System was analyzed in the Chinese Gelao minority group of 234 unrelated males living in Daozhen Gelao-Miao Autonomous County in Guizhou Province, southwest China. A total of 216 different haplotypes were detected, of which 199 haplotypes were unique. The overall haplotype diversity (HD) and discrimination capacity (DC) were 0.9996 and 0.9231, respectively. The gene diversity (GD) ranged from 0.4159 (DYS438) to 0.9650 (DYS385a/b). The haplotype frequencies varied from 0.0043 to 0.0128. The population data presented here showed high genetic polymorphism and extraordinary discriminatory power in the studied population. Population difference was observed between the Chinese Gelao ethnicity and 42 populations out of overall 59 neighboring populations in Asia region. Both multidimensional scaling (MDS) and the phylogenetic tree demonstrated that the genetic structure affinity and differentiation with Chinese Gelao ethnicity were identified in those populations geographically adjacent (Hunan Han) and distant (Chinese Tibetan), respectively. In conclusion, our study enriched the Chinese ethnic genetic information and could be used as a powerful tool in forensic genetics for male testing, paternal lineage analysis, and Gelao ethnic population evolutionary studies.
Similar content being viewed by others
References
Gao T, Yun L, Gao S, Gu Y, He W, Luo H, Hou Y (2016) Population genetics of 23 Y-STR loci in the Mongolian minority population in Inner Mongolia of China. Int J Legal Med 130(6):1509–1511. https://doi.org/10.1007/s00414-016-1433-1
Purps J, Siegert S, Willuweit S, Nagy M, Alves C, Salazar R, Angustia SMT, Santos LH, Anslinger K, Bayer B, Ayub Q, Wei W, Xue Y, Tyler-Smith C, Bafalluy MB, Martínez-Jarreta B, Egyed B, Balitzki B, Tschumi S, Ballard D, Court DS, Barrantes X, Bäßler G, Wiest T, Berger B, Niederstätter H, Parson W, Davis C, Budowle B, Burri H, Borer U, Koller C, Carvalho EF, Domingues PM, Chamoun WT, Coble MD, Hill CR, Corach D, Caputo M, D’Amato ME, Davison S, Decorte R, Larmuseau MHD, Ottoni C, Rickards O, Lu D, Jiang C, Dobosz T, Jonkisz A, Frank WE, Furac I, Gehrig C, Castella V, Grskovic B, Haas C, Wobst J, Hadzic G, Drobnic K, Honda K, Hou Y, Zhou D, Li Y, Hu S, Chen S, Immel UD, Lessig R, Jakovski Z, Ilievska T, Klann AE, García CC, de Knijff P, Kraaijenbrink T, Kondili A, Miniati P, Vouropoulou M, Kovacevic L, Marjanovic D, Lindner I, Mansour I, al-Azem M, Andari AE, Marino M, Furfuro S, Locarno L, Martín P, Luque GM, Alonso A, Miranda LS, Moreira H, Mizuno N, Iwashima Y, Neto RSM, Nogueira TLS, Silva R, Nastainczyk-Wulf M, Edelmann J, Kohl M, Nie S, Wang X, Cheng B, Núñez C, Pancorbo MM, Olofsson JK, Morling N, Onofri V, Tagliabracci A, Pamjav H, Volgyi A, Barany G, Pawlowski R, Maciejewska A, Pelotti S, Pepinski W, Abreu-Glowacka M, Phillips C, Cárdenas J, Rey-Gonzalez D, Salas A, Brisighelli F, Capelli C, Toscanini U, Piccinini A, Piglionica M, Baldassarra SL, Ploski R, Konarzewska M, Jastrzebska E, Robino C, Sajantila A, Palo JU, Guevara E, Salvador J, Ungria MCD, Rodriguez JJR, Schmidt U, Schlauderer N, Saukko P, Schneider PM, Sirker M, Shin KJ, Oh YN, Skitsa I, Ampati A, Smith TG, Calvit LS, Stenzl V, Capal T, Tillmar A, Nilsson H, Turrina S, de Leo D, Verzeletti A, Cortellini V, Wetton JH, Gwynne GM, Jobling MA, Whittle MR, Sumita DR, Wolańska-Nowak P, Yong RYY, Krawczak M, Nothnagel M, Roewer L (2014) A global analysis of Y-chromosomal haplotype diversity for 23 STR loci. Forensic Sci Int Genet 12:12–23. https://doi.org/10.1016/j.fsigen.2014.04.008
Li CT, Ma T, Zhao SM et al (2011) Development of 11 X-STR loci typing system and genetic analysis in Tibetan and Northern Han populations from China. Int J Legal Med 125(5):753–756. https://doi.org/10.1007/s00414-011-0592-3
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215. https://doi.org/10.1093/nar/16.3.1215
Gusmao L, Butler JM, Carracedo A et al (2006) DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int 157(2–3):187–197. https://doi.org/10.1016/j.forsciint.2005.04.002
He G, Chen P, Zou X et al (2017) Genetic polymorphism investigation of the Chinese Yi minority using PowerPlex(R) Y23 STR amplification system. Int J Legal Med 131(3):663–666. https://doi.org/10.1007/s00414-017-1537-2
Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, de Knijff P, Jobling MA (2002) High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci Int 125(1):42–51. https://doi.org/10.1016/S0379-0738(01)00627-2
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. https://doi.org/10.1093/molbev/msm092
Acknowledgements
We would like to acknowledge all volunteers who contributed samples for this study.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81401562) and the PhD Scientific Research Start-up Fund of Affiliated Hospital of Zunyi Medical University (No. 201501).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Informed consent was obtained before sample collection. The study reported in this paper was approved by the Ethics Committee of Zunyi Medical University.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chen, P., Han, Y., He, G. et al. Genetic diversity and phylogenetic study of the Chinese Gelao ethnic minority via 23 Y-STR loci. Int J Legal Med 132, 1093–1096 (2018). https://doi.org/10.1007/s00414-017-1743-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-017-1743-y