Abstract
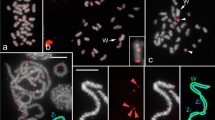
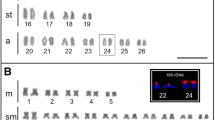
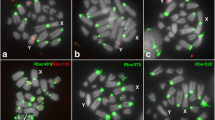
To better understand the structure and variability of the 45S rDNA cistron and its evolutionary dynamics in grasshoppers, we performed a detailed analysis combining classical and molecular cytogenetic data with whole-genome sequencing in Abracris flavolienata, which shows extraordinary variability in the chromosomal distribution for this element. We found astonishing variability in the number and size of rDNA clusters at intra- and inter-population levels. Interestingly, FISH using distinct parts of 45S rDNA cistron (18S rDNA, 28S rDNA, and ITS1) as probes revealed a distinct number of clusters, suggesting independent mobility and amplification of the 45S rDNA components. This hypothesis is consistent with the higher genomic coverage of almost the entire cistron of 45S rDNA observed in A. flavolineata compared to other grasshoppers, besides coverage variability along the 45S rDNA cistron in the species. In addition, these differences in coverage for distinct components of the 45S rDNA cistron indicate emergence of pseudogenes evidenced by existence of truncated sequences, demonstrating the rDNA dynamics in the species. Although the chromosomal distribution of 18S rDNA was highly variable, the chromosomes 1, 3, 6, and 9 harbored rDNA clusters in all individuals with the occurrence of NOR activity in pair 9, suggesting ancestry or selective pressures to prevent pseudogenization of rDNA sequences in this chromosome pair. Additionally, small NORs and cryptic rDNA loci were observed. Finally, there was no evidence of enrichment and association of transposable elements, at least, inside or nearby rDNA cistron. These findings broaden our knowledge of rDNA dynamics, revealing an independent movement and amplification of segments of 45S rDNA cistron, which in A. flavolineata could be attributed to ectopic recombination.





Similar content being viewed by others
References
Bi K, Bogart JP, Fu J (2009) A Populational survey of 45S rDNA polymosphism in the Jefferson salamander Ambystoma jeffersonianum revealed by fluorescence in situ hybridzation (FISH). Curr Zool 55:145–149
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bueno D, Palacios-Gimenez OM, Cabral-De-Mello DC (2013) Chromosomal mapping of repetitive DNAs in the grasshopper Abracris flavolineata reveal possible ancestry of the B chromosome and H3 histone spreading. PLoS One 8:e66532. https://doi.org/10.1371/journal.pone.0066532
Cabral-de-Mello DC, Moura RC, Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and a complement. Heredity 104:393–400. https://doi.org/10.1038/hdy.2009.126
Cabrero J, Camacho JPM (2008) Location and expression of ribosomal RNA genes in grasshoppers: abundance of silent and cryptic loci. Chromosom Res 16:595–607. https://doi.org/10.1007/s10577-008-1214-x
Cabrero J, Perfectti F, Gómez R, Camacho JPM, Lopéz-Leon MD (2003) Population variation in the a chromosome distribution of satellite DNA and ribosomal DNA in the grasshopper Eyprepocnemis plorans. Chromosom Res 11:375–381. https://doi.org/10.1023/A:1024127525756
Cai Q, Zhang DM, Liu ZL, Wang XR (2006) Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann Bot 97:715–722. https://doi.org/10.1093/aob/mcl030
Castro J, Rodriâguez S, Pardo BG, Sánchez L, Martínez P (2001) Population analysis of an unusual NOR-site polymorphism in brown trout (Salmo trutta L.). Heredity 86:291–302
Cella DM, Ferreira A (1991) The cytogenetics of Abracris flavolineata (Orthoptera, Caelifera, Ommatolampinae, Abracrini). Rev Bras Genet 14:315–329
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220. https://doi.org/10.1038/371215a0
Datson PM, Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosom Res 14:846–857. https://doi.org/10.1007/s10577-006-1092-z
Dover GA (1986) Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet 2:159–165. https://doi.org/10.1016/0168-9525(86)90211-8
Dubcovsky J, Dvorák J (1995) Ribosomal RNA multigene loci - nomads of the Triticeae genomes. Genetics 140:1367–1377
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175:477–485. https://doi.org/10.1534/genetics.107.071399
Eirín-López JM, Rebordinos L, Rooney AP, Rozas J (2012) The birth-and-death evolution of multigene families revisited. Genome Dyn 7:170–196. https://doi.org/10.1159/000337119
Garcia S, Garnatje T, Kovařík A (2012) Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121:389–394. https://doi.org/10.1007/s00412-012-0368-7
Garcia S, Kovařík A, Leitch AR, Garnatie T (2017) Cytogenetic features of rRNA genes across land plants: analysis of the plant rDNA database. Plant J 89:1020–1030. https://doi.org/10.1111/tpj.13442
Garrido-Ramos MA (2017) Satellite DNA: an evolving topic. Genes 8:230. https://doi.org/10.3390/genes8090230
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gordon A, Hannon GJ (2010) Fastx-toolkit. FASTQ/a short-reads preprocessing tools. Available at http://hannonlab.cshl.edu/fastx_toolkit
Gornung E (2013) Twenty years of physical mapping of major ribosomal RNA genes across the Teleosts: a review of research. Cytogenet Genome Res 141:90–102. https://doi.org/10.1159/000354832
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res 41:129–129. https://doi.org/10.1093/nar/gkt371
Han JS (2010) Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA 1:15. https://doi.org/10.1186/1759-8753-1-15
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877. https://doi.org/10.1101/gr.9.9.868
Jo S-H, Koo D-H, Kim JF, Hur C-G, Lee S, Yang T-J, Know S-Y, Choi D (2009) Evolution of ribosomal DNA-derived satellite repeat in tomato genome. BMC Plant Biol 9:42. https://doi.org/10.1186/1471-2229-9-42
Kent WJ (2002) BLAT-the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lohe AR, Roberts PA (1990) An unusual Y chromosome of Drosophila simulans carrying amplified rDNA spacer without rRNA genes. Genetics 125:399–406
Maggini F, Cremonini R, Zolfino C, Tucci GF, D'Ovidio R, Delre V, DePace D, Scarascia Mugnozza GT, Cionini PG (1991) Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma 100:229–234. https://doi.org/10.1007/BF00344156
Mallatt J, Giribet G (2006) Further use of nearly complete 28S and 18S rRNA genes to classify Ecdysozoa: 37 more arthropods and a kinorhynchn. Mol Phylogenet Evol 40:772–794. https://doi.org/10.1016/j.ympev.2006.04.021
Mantovani M, dos Santos LD, Moreira-Filho O (2005) Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123:221–216. https://doi.org/10.1007/s10709-004-2281-3
Montgomery EA, Huang SM, Langley CH, Judd BH (1991) Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129:1085–1098
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. https://doi.org/10.1146/annurev.genet.39.073003.112240
Nguyen P, Sahara K, Yoshido A, Marec F (2010) Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica 138:343–354. https://doi.org/10.1007/s10709-009-9424-5
Pedrosa-Harand A, de Almeida CC, Mosiolek M, Blair MW, Schweizer D, Guerra M (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112:924–933. https://doi.org/10.1007/s00122-005-0196-8
Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE (2003) Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol 20:880–892. https://doi.org/10.1093/molbev/msg102
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A 83:2934–2938. https://doi.org/10.1073/pnas.83.9.2934
Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosom Res 12:153–161. https://doi.org/10.1023/B:CHRO.0000013168.61359.43
Reed KM, Phillips RB (2000) Structure and organization of the rDNA intergenic spacer in lake trout (Salvelinus namaycush). Chromosom Res 8:5–16. https://doi.org/10.1023/A:1009214800251
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225. https://doi.org/10.1186/1471-2148-12-225
Rocha MF, Melo NF, Souza MJ (2011) Comparative cytogenetic analysis of two grasshopper species of the tribe Abracrini (Ommatolampinae, Acrididae). Gent Mol Biol 34:214–219. https://doi.org/10.1590/S1415-47572011000200008
Rufas JS, Gimenez-Abian J, Suja JA, Garcia De La Vega C (1987) Chromosome organization in meiosis revealed by light microscope analysis of silver-stained cores. Genome 29:706–712. https://doi.org/10.1139/g87-121
Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM (2016) High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci Rep 6:28333. https://doi.org/10.1038/srep28333
Ruiz-Ruano FJ, Cabrero J, López-León MD, Camacho JPM (2017) Satellite DNA content illuminates the ancestry of a supernumerary (B) chromosome. Chromosoma 126:487–500. https://doi.org/10.1007/s00412-016-0611-8
Ruiz-Ruano FJ, Castillo-Martínez J, Cabrero J, Gómez R, Camacho JPM, López-León MD (2018) High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 127:323–340. https://doi.org/10.1007/s00412-018-0666-9
Schmid M, Steinlein C, Feichtinger W, Nanda I (2017) Chromosome banding in amphibia. XXXV. Highly mobile nucleolus organizing regions in Craugastor fitzingeri (Anura, Craugastoridae). Cytogenet Genome Res 152:180–193. https://doi.org/10.1159/000481554
Schubert I (1984) Mobile nucleolus organizing regions (NORs) in Allium (Liliaceae s. lat.) - inferences from the specifity of silver staining. Plant Syst Evol 144:291–305. https://doi.org/10.1007/BF00984139
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148. https://doi.org/10.1007/BF00328466
Shishido R, Sano Y, Fukui K (2000) Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol Gen Genet 263:586–591
Smit AFA, Hubley R, Green P (2017) RepeatMasker Open-4.0. Available at http://www.repeatmasker.org
Sochorová J, Garcia S, Gálvez F, Symonová R, Kovarik A (2018) Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database. Chromosoma 127:141–150. https://doi.org/10.1007/s00412-017-0651-8
Stupar RM, Song J, Tek AL, Cheng Z, Dong F, Jiang J (2002) Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162:1435–1444
Symonová R, Majtánová Z, Sember A, Staaks GBO, Bohlen J, Freyhof J, Rábová M, Ráb P (2013) Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol Biol 13:42. https://doi.org/10.1186/1471-2148-13-42
Teruel M, Cabrero J, Perfectti F, Camacho JMP (2010) B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 119:217–225. https://doi.org/10.1007/s00412-009-0251-3
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Veltsos P, Keller I, Nichols RA (2009) Geographically localised bursts of ribosomal DNA mobility in the grasshopper Podisma pedestris. Heredity 103:54–61. https://doi.org/10.1038/hdy.2009.32
Webb GC, White MJD, Contreras N, Cheney J (1978) Cytogenetics of the parthenogenetic grasshopper Warramaba (formely Moraba) virgo and its bisexual relatives. IV. Chromosome banding studies. Chromosoma 67:309–339. https://doi.org/10.1007/BF00285964
Acknowledgements
The authors are grateful to Dr. Frantisek Marec for critical reading of an earlier version of the manuscript, to the anonymous reviewers for valuable suggestions and to Dr. Edison Zefa for providing samples from Murici/AL.
Funding
This study was partially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (process number 2015/16661-1 and 2014/11763-8), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). DCCM received a research productivity fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process number 305300/2017-2), and Ana Beatriz Stein Machado Ferretti received a scientific initiation scholarship from PIBIC-UNESP, CNPq.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Material 1
Map showing the geographical location for each population studied. (JPG 245 kb)
Supplementary Material 2
Contigs containing sequences associated with the 45S rDNA, annotation of associated sequence, and frequency of association. (DOCX 70 kb)
Supplementary Material 3
Difference in size for classification of large (L) and small (S) sites of 18S rDNA in chromosomes of Abracris flavolineata, (a) meiotic chromosomes (metaphase I) and (b) mitotic chromosomes from embryo. The large signals (L) occurred in the pericentromeric region extending to the short chromosomal arm, while the small signals (S) appeared as small dots in the short arms near to the cetromeric region. (JPG 657 kb)
Supplementary Material 4
Occurrence of large (L) and small (S) 18S rDNA clusters and their frequency in individual chromosomes of Abracris flavolineata for the six populations studied. Asterisks indicate heteromorphism (monosomicity). (DOCX 43 kb)
Supplementary Material 5
FISH mapping using the probe ITS1–2 in individuals from six populations of Abracris flavolineata studied. (JPG 2147 kb)
Rights and permissions
About this article
Cite this article
Ferretti, A.B.S.M., Ruiz-Ruano, F.J., Milani, D. et al. How dynamic could be the 45S rDNA cistron? An intriguing variability in a grasshopper species revealed by integration of chromosomal and genomic data. Chromosoma 128, 165–175 (2019). https://doi.org/10.1007/s00412-019-00706-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00706-8