Abstract
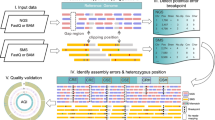
The complexity of eukaryote genomes makes assembly errors inevitable in the process of constructing reference genomes. Next-generation sequencing (NGS) could provide an efficient way to validate previously assembled genomes. Here, we exploited NGS data to interrogate the chicken reference genome and identified 35 pairs of nearly identical regions with >99.5 % sequence similarity and a median size of 109 kb. Several lines of evidence, including read depth, the composition of junction sequences, and sequence similarity, suggest that these regions present genome assembly errors and should be excluded from forthcoming genomic studies.


Similar content being viewed by others
References
Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297(5583):1003–1007
Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW (2003) Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 4(4):R25
Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM (2013) Ensembl 2013. Nucleic Acids Res 41:D48–55, Database issue
Kelley DR, Salzberg SL (2010) Detection and correction of false segmental duplications caused by genome mis-assembly. Genome Biol 11(3):R28
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12(4):656–664
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Myers EW (1995) Toward simplifying and accurately formulating fragment assembly. J Comput Biol 2(2):275–290
Phillippy AM, Schatz MC, Pop M (2008) Genome assembly forensics: finding the elusive mis-assembly. Genome Biol 9(3):R55
Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, Hallbook F, Besnier F, Carlborg O, Bed’hom B, Tixier-Boichard M, Jensen P, Siegel P, Lindblad-Toh K, Andersson L (2010) Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464(7288):587–591
Salzberg SL, Yorke JA (2005) Beware of mis-assembled genomes. Bioinformatics 21(24):4320–4321
Yoon S, Xuan Z, Makarov V, Ye K, Sebat J (2009) Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 19(9):1586–1592
Acknowledgments
QZ was supported by the Department of Human Evolutionary Biology, Harvard University. NB acknowledges postdoctoral research funding from the Swedish Research Council (VR grant 2009-693). We thank the anonymous reviewers for the helpful comments on an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 49 kb)
Rights and permissions
About this article
Cite this article
Zhang, Q., Backström, N. Assembly errors cause false tandem duplicate regions in the chicken (Gallus gallus) genome sequence. Chromosoma 123, 165–168 (2014). https://doi.org/10.1007/s00412-013-0443-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-013-0443-8