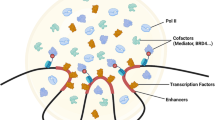
Abstract
The assembly of the transcription machinery is a key step in gene activation, but even basic details of this process remain unclear. Here we discuss the apparent discrepancy between the classic sequential assembly model based mostly on biochemistry and an emerging dynamic assembly model based mostly on fluorescence microscopy. The former model favors a stable transcription complex with subunits that cooperatively assemble in order, whereas the latter model favors an unstable complex with subunits that may assemble more randomly. To confront this apparent discrepancy, we review the merits and drawbacks of the different experimental approaches and list potential biasing factors that could be responsible for the different interpretations of assembly. We then discuss how these biases might be overcome in the future with improved experiments or new techniques. Finally, we discuss how kinetic models for assembly may help resolve the ordered and stable vs. random and dynamic assembly debate.



Similar content being viewed by others
Abbreviations
- PIC:
-
Preinitiation complex
- ChIP:
-
Chromatin immunoprecipitation
- GFP:
-
Green fluorescent protein
- FRAP:
-
Fluorescence recovery after photobleaching
- FLIP:
-
Fluorescence loss in photobleaching
- FCS:
-
Fluorescence correlation spectroscopy
- SMT:
-
Single-molecule tracking
- Fabs:
-
Antigen binding fragments
- FCCS:
-
Fluorescence cross-correlation spectroscopy
- MFPT:
-
Mean first passage time
- SPM:
-
Surface plasmon resonance
- TIRFM:
-
Total internal reflection fluorescence microscopy
References
Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667–678
Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S (1994) Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev 8:1920–1934
Bonham AJ, Neumann T, Tirrell M, Reich NO (2009) Tracking transcription factor complexes on DNA using total internal reflectance fluorescence protein binding microarrays. Nucleic Acids Res 37:e94–e94
Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G (2006) A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-κB-dependent gene activity. EMBO J 25:798–810
Buratowski S (1994) The basics of basal transcription by RNA polymerase II. Cell 77:1–3
Buratowski S (2000) Snapshots of RNA polymerase II transcription initiation. Curr Opin Cell Biol 12:320–325
Bushnell DA, Bamdad C, Kornberg RD (1996) A minimal set of RNA polymerase II transcription protein interactions. J Biol Chem 271:20170–20174
Catez F, Ueda T, Bustin M (2006) Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol 13:305–310
Chen D, Hinkley CS, Henry RW, Huang S (2002) TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 13:276–284
Collins GA, Tansey WP (2006) The proteasome: a utility tool for transcription? Curr Opin Genet Dev 16:197–202
Conaway RC, Conaway JW (1993) General initiation factors for RNA polymerase II. Annu Rev Biochem 62:161–190
Cook PR (2001) Principles of nuclear structure and function, 1st edn. Wiley-Liss, New York
D'Alessio JA, Wright KJ, Tjian R (2009) Shifting players and paradigms in cell-specific transcription. Mol Cell 36:924–931
Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH (2007) In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14:796–806
Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, Tjian R, Singer RH (2009) Imaging transcription in living cells. Annu Rev Biophys 38:173–196
Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP (2008) Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci USA 105:19649–19654
de Graaf P, Mousson F, Geverts B, Scheer E, Tora L, Houtsmuller AB, Timmers HTM (2010) Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. J Cell Sci 123:2663–2671
Deng W, Roberts SGE (2007) TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 116:417–429
Dezwaan DC, Freeman BC (2008) HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle 7:1006–1012
Dinant C, Luijsterburg MS, Höfer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, van Driel R (2009) Assembly of multiprotein complexes that control genome function. J Cell Biol 185:21–26
D'Orsogna MR, Chou T (2005) First passage and cooperativity of queuing kinetics. Phys Rev Lett 95:170603
Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623–1626
Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL, DeFranco DB (2004) Molecular chaperones function as steroid receptor nuclear mobility factors. PNAS 101:2876–2881
Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C (2009) A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA 106:20240–20245
Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford R, Warren BS, Hager GL (2002) ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol 22:3255–3263
Freeman BC, Yamamoto KR (2002) Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232–2235
Giglia-Mari G, Theil AF, Mari P-O, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, Fousteri M, Hoeijmakers JHJ, Vermeulen W (2009) Differentiation driven changes in the dynamic organization of basal transcription initiation. PLoS Biol 7:e1000220
Gorski SA, Snyder SK, John S, Grummt I, Misteli T (2008) Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell 30:486–497
Guermah M, Kim J, Roeder RG (2009) Transcription of in vitro assembled chromatin templates in a highly purified RNA polymerase II system. Methods 48:353–360
Hager GL, Elbi C, Johnson TA, Voss T, Nagaich AK, Schiltz RL, Qiu Y, John S (2006) Chromatin dynamics and the evolution of alternate promoter states. Chromosome Res 14:107–116
Hager GL, McNally JG, Misteli T (2009) Transcription dynamics. Mol Cell 35:741–753
Hahn S (2004) Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol 11:394–403
Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369–379
Hayashi-Takanaka Y, Yamagata K, Nozaki N, Kimura H (2009) Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. J Cell Biol 187:781–790
Hemmerich P, Schmiedeberg L, Diekmann S (2010) Dynamic as well as stable protein interactions contribute to genome function and maintenance. Chromosome Res 19:131–151
Hoogstraten D, Nigg AL, Heath H, Mullenders LHF, van Driel R, Hoeijmakers JHJ, Vermeulen W, Houtsmuller AB (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell 10:1163–1174
Hoopes BC, LeBlanc JF, Hawley DK (1992) Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem 267:11539–11547
Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA 71:4135–4139
Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ (2011) Ordered and dynamic assembly of single spliceosomes. Science 331:1289–1295
Iwahara J, Clore GM (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440:1227–1230
Johnson TA, Elbi C, Parekh BS, Hager GL, John S (2008) Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell 19:3308–3322
Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT (2008) The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20:253–259
Kadonaga JT (1990) Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J Biol Chem 265:2624–2631
Kaiser TE, Intine RV, Dundr M (2008) De novo formation of a subnuclear body. Science 322:1713–1717
Karpova TS, Chen TY, Sprague BL, McNally JG (2004) Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep 5:1064–1070
Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG (2008) Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319:466–469
Kimura H, Tao Y, Roeder RG, Cook PR (1999) Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol 19:5383–5392
Kimura H, Sugaya K, Cook PR (2002) The transcription cycle of RNA polymerase II in living cells. J Cell Biol 159:777–782
Kornberg RD (2007) The molecular basis of eukaryotic transcription. Proc Natl Acad Sci USA 104:12955–12961
Kuipers MA, Stasevich TJ, Sasaki T, Wilson KA, Hazelwood KL, McNally JG, Davidson MW, Gilbert DM (2011) Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J Cell Biol 192:29–41
Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14:2551–2569
Li G, Margueron R, Hu G, Stokes D, Wang Y-H, Reinberg D (2010) Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell 38:41–53
Lieberman BA, Nordeen SK (1997) DNA intersegment transfer, how steroid receptors search for a target site. J Biol Chem 272:1061–1068
Linnell J, Mott R, Field S, Kwiatkowski DP, Ragoussis J, Udalova IA (2004) Quantitative high-throughput analysis of transcription factor binding specificities. Nucleic Acids Res 32:e44–e44
Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Moné MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Höfer T (2010) Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol 189:445–463
Manley JL, Fire A, Cano A, Sharp PA, Gefter ML (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA 77:3855–3859
Martin BR, Giepmans BNG, Adams SR, Tsien RY (2005) Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol 23:1308–1314
McNairn AJ, Okuno Y, Misteli T, Gilbert DM (2005) Chinese hamster ORC subunits dynamically associate with chromatin throughout the cell-cycle. Exp Cell Res 308:345–356
Mizushima S, Nomura M (1970) Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226:1214
Mu D, Hsu DS, Sancar A (1996) Reaction mechanism of human DNA repair excision nuclease. J Biol Chem 271:8285–8294
Mueller F, Wach P, McNally JG (2008) Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J 94:3323–3339
Mueller F, Mazza D, Stasevich TJ, McNally JG (2010) FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol 22:403–411
Muller F, Tora L (2004) The multicoloured world of promoter recognition complexes. EMBO J 23:2–8
Murthy KPN, Kehr KW (1989) Mean first-passage time of random walks on a random lattice. Phys Rev A 40:2082
Murthy KPN, Kehr KW (1990) Erratum: mean first-passage time of random walks on a random lattice. Phys Rev A 41:1160
Nagaich AK, Walker DA, Wolford R, Hager GL (2004) Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14:163–174
Nalley K, Johnston SA, Kodadek T (2006) Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442:1054–1057
Nalley K, Johnston SA, Kodadek T (2009) Nalley et al. reply. Nature 461:E8
Perlmann T, Eriksson P, Wrange O (1990) Quantitative analysis of the glucocorticoid receptor-DNA interaction at the mouse mammary tumor virus glucocorticoid response element. J Biol Chem 265:17222–17229
Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM (2010) Single cell analysis of transcriptional activation dynamics. PLoS One 5:e10272
Roberts DN, Stewart AJ, Huff JT, Cairns BR (2003) The RNA polymerase III transcriptome revealed by genome-wide localization and activity–occupancy relationships. Proc Natl Acad Sci USA 100:14695–14700
Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21:327–335
Santoso B, Kadonaga JT (2006) Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat Struct Mol Biol 13:131–139
Schneider DA, Nomura M (2004) RNA polymerase I remains intact without subunit exchange through multiple rounds of transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101:15112–15117
Seki T, Diffley JFX (2000) Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA 97:14115–14120
Shumaker-Parry JS, Aebersold R, Campbell CT (2004) Parallel, quantitative measurement of protein binding to a 120-element double-stranded DNA array in real time using surface plasmon resonance microscopy. Anal Chem 76:2071–2082
Siebrasse JP, Kubitscheck U (2009) Single molecule tracking for studying nucleocytoplasmic transport and intranuclear dynamics. Methods Mol Biol 464:343–361
Sikorski TW, Buratowski S (2009) The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 21:344–351
Sinnecker D, Voigt P, Hellwig N, Schaefer M (2005) Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry 44:7085–7094
Sisan DR, Yarar D, Waterman CM, Urbach JS (2010) Event ordering in live-cell imaging determined from temporal cross-correlation asymmetry. Biophys J 98:2432–2441
Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT (2008) Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci USA 105:13304–13308
Stasevich TJ, Mueller F, Michelman-Ribeiro A, Rosales T, Knutson JR, McNally JG (2010) Cross-validating FRAP and FCS to quantify the impact of photobleaching on in vivo binding estimates. Biophys J 99:3093–3101
Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG (2004) Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol 24:2682–2697
Sung M-H, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL, Bianchi ME, Agresti A (2009) Sustained oscillations of NF-κB produce distinct genome scanning and gene expression profiles. PLoS One 4:e7163
Talkington MWT, Siuzdak G, Williamson JR (2005) An assembly landscape for the 30S ribosomal subunit. Nature 438:628–632
Tokunaga M, Imamoto N, Sakata-Sogawa K (2008) Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5:159–161
Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-Suárez M, Puthenveetil S, Ting AY (2010) A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci USA 107:10914–10919
van Werven FJ, van Teeffelen HAAM, Holstege FCP, Timmers HTM (2009) Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16:1043–1048
Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P (2008) Dynamics of component exchange at PML nuclear bodies. J Cell Sci 121:2731–2743
Weil PA, Luse DS, Segall J, Roeder RG (1979) Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell 18:469–484
Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PIH, Lygerou Z (2007) Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J 26:1303–1314
Yao J, Munson KM, Webb WW, Lis JT (2006) Dynamics of heat shock factor association with native gene loci in living cells. Nature 442:1050–1053
Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT (2007) Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28:978–990
Yean D, Gralla J (1997) Transcription reinitiation rate: a special role for the TATA box. Mol Cell Biol 17:3809–3816
Yudkovsky N, Ranish JA, Hahn S (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225–229
Zawel L, Kumar KP, Reinberg D (1995) Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev 9:1479–1490
Zobeck KL, Buckley MS, Zipfel WR, Lis JT (2010) Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40:965–975
Acknowledgments
We would like to thank Hiroshi Kimura, David Auble, Akhilesh Nagaich, and Avin Lalmansingh for the critical reading of the manuscript and helpful comments. This research was supported in part by the intramural program of the National Institutes of Health, National Cancer Institute, and the Center for Cancer Research. TJS was also supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Foreign Researchers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Erich Nigg
Rights and permissions
About this article
Cite this article
Stasevich, T.J., McNally, J.G. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both?. Chromosoma 120, 533–545 (2011). https://doi.org/10.1007/s00412-011-0340-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-011-0340-y