Abstract
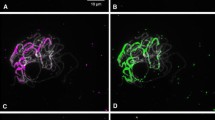
We sequenced a continuous 326-kb DNA stretch of a microscopically defined centromeric region of tomato chromosome 12. A total of 84% of the sequence (270 kb) was composed of a nested complex of repeat sequences including 27 retrotransposons, two transposable elements, three MITEs, two terminal repeat retrotransposons in miniature (TRIMs), ten unclassified repeats and three chloroplast DNA insertions. The retrotransposons were grouped into three families of Ty3-Gypsy type long terminal repeat (LTR) retrotransposons (PCRT1–PCRT3) and one LINE-like retrotransposon (PCRT4). High-resolution fluorescence in situ hybridization analyses on pachytene complements revealed that PCRT1a occurs on the pericentromere heterochromatin blocks. PCRT1 was the prevalent retrotransposon family occupying more than 60% of the 326-kb sequence with 19 members grouped into eight subfamilies (PCRT1a–PCRT1h) based on LTR sequence. The PCRT1a subfamily is a rapidly amplified element occupying tens of megabases. The other PCRT1 subfamilies (PCRT1b–PCRT1h) were highly degenerated and interrupted by insertions of other elements. The PCRT1 family shows identity with a previously identified tomato-specific repeat TGR2 and a CENP-B like sequence. A second previously described genomic repeat, TGR3, was identified as a part of the LTR sequence of an Athila-like PCRT2 element of which four copies were found in the 326-kb stretch. A large block of trinucleotide microsatellite (CAA)n occupies the centromere and large portions of the flanking pericentromere heterochromatin blocks of chromosome 12 and most of the other chromosomes. Five putative genes in the remaining 14% of the centromere region were identified, of which one is similar to a transcription regulator (ToCPL1) and a candidate jointless-2 gene.









Similar content being viewed by others
References
Ananiev EV, Phillips RL, Rines HW (1998) Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci U S A 95:13073–13078
Barton DW (1950) Pachytene morphology of the tomato chromosome complement. Am J Bot 37:639–643
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Broun P, Tanksley SD (1996) Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol Gen Genet 250:39–49
Budiman MA, Chang SB, Lee S, Yang TJ, Zhang HB, de Jong JH, Wing RA (2004) Localization of jointless-2 gene in the centromeric region of tomato chromosome 12 based on high resolution genetic and physical mapping. Theor Appl Genet 108:190–196
Cheng Z, Murata M (2003) A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164:665–672
Cheng Z, Presting GG, Buell CR, Wing RA, Jiang J (2001) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157:1749–1757
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Clarke L (1998) Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev 8:212–218
Copenhaver GP, Nckel K, Kuromori T, Benito M, Kaul S, Lin X, Bvan M, Murphy G, Harris B, Parnell LD, McCombie WR, Martienssen RA, Marra M, Preuss D (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286:2468–2474
Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci U S A 95:8135–8140
Entani T, Iwano M, Shiba H, Takayama S (1999) Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theor Appl Genet 99:391–397
Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res 8:175–185
Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X et al (2002) Sequence and analysis of rice chromosome 4. Nature 420:316–320
Frary A, Presting GG, Tanksley S (1996) Molecular mapping of the centromeres of tomato chromosomes 7 and 9. Mol Gen Genet 250:295–304
Fukui KN, Suzuki G, Lagudah ES, Rahman S, Appels R, Yamamoto M, Mukai Y (2001) Physical arrangement of retrotransposon-related repeats in centromeric regions of wheat. Plant Cell Physiol 42:189–196
Ganal MW, Lapitan N, Tanksley SD (1991) Macrostructure of the tomato telomeres. Plant Cell 3:87–94
Ganal MW, Broun P, Tanksley SD (1992) Genetic mapping of tandemly repeated telomeric DNA sequences in tomato (Lycopersicon esculentum). Genomics 14:444–448
Gindullis F, Desel C, Galasso I, Schdidt T (2001) The large-scale organization of the centromeric region in Beta species. Genome Res 11:253–265
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Guy J, Hearn T, Crosier M, Mudge J, Viggiano L, Koczan D, Thiesen HJ, Bailey JA, Horvath JE, Eichler EE, Earthrowl ME, Deloukas P, French L, Rogers J, Benley D, Jackson MS (2003) Genomic sequence and transcriptional profile of the boundary between pericentromere satellites and genes on human chromosome arm 10p. Genome Res 13:159–172
Harrison GE, Heslop-Harrison JS (1995) Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet 90:157–165
Henikoff S (2002) Near the edge of a chromosome’s black hole. Trends Genet 18:165–167
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Henning KA, Novotny EA, Compton ST, Guan XY, Liu PP, Ashlock MA (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc Natl Acad Sci U S A 96:592–597
Hudakova S, Michalek W, Presting GG, ten Hoopen R, dos Santos K, Jasencakova Z, Schubert I (2001) Sequence organization of barley centromeres. Nucleic Acids Res 29:5029–5035
Jiang N, Wessler SR (2001) Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13:2553–2564
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16:571–581
Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, Sharkhuu A, Manabe Y, Yokoi S, Zhu JK, Bressan RA, Hasegawa PM (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci U S A 99:10893–10898
Ku HM, Vision T, Liu J, Tanksley SD (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci U S A 97:9121–9126
Kulikova O, Geurts R, Lamine M, Kim DJ, Cook DR, Leunissen J, de Jong JH, Roe BA, Bisseling T (2004) Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. Chromosoma 113:276–283
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2000) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res 7:315–321
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res 8:285–290
Kurata N, Nonomura KI, Harushima Y (2002) Rice genome organization: the centromere and genome interactions. Ann Bot 90:427–435
Lafarge S, Montané MH (2003) Caracterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res 31:1148–1155
Lamb JC, Birchler JA (2003) The role of DNA sequence in centromere formation. Genome Biol 4:214
Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406:910–913
Mao L, Begum D, Goff SA, Wing RA (2001) Sequence and analysis of the tomato JOINTLESS locus. Plant Physiol 126:1331–1340
Nagaki K, Song J, Stupar M, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, Buell CR, Jiang J (2003a) Molecular and cytological analysis of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163:759–770
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J (2003b) Chromatin immunoprecipitation reveals that the 180 bp satellite repeat is the key functional DNA element of Arabidopsis thanlina centromeres. Genetics 163:1221–1225
Page BT, Wanous MK, Birchler JA (2001) Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159:291–302
Parsons JD (1995) Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci 11:615–619
Presting GG, Frary A, Pillen K, Tanksley SD (1996) Telomere-homologous sequences occur near the centromeres of many tomato chromosomes. Mol Gen Genet 251:526–531
Rossberg M, Theres K, Acarkan A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R (2001) Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13:979–988
Round EK, Flowers SK, Richards EJ (1997) Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res 7:1045–1053
Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586
Stegemann S, Hartmann S, Ruf S, Bock R (2003) High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci U S A 100:8828–8833
Sun X, Wahlstrom J, Karpen G (1997) Molecular structure of a functional Drosophila centromere. Cell 91:1007–1019
Sun X, Le HD, Wahlstrom JM, Karpen GH (2003) Sequence analysis of a functional Drosophila centromere. Genome Res 13:182–194
Thompson H, Schmidt R, Brandes A, Heslop-Harrison JS, Dean C (1996) A novel repetitive sequence associated with the centromeric regions of Arabidopsis thaliana chromosomes. Mol Gen Genet 253:247–252
van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14:1441–1456
van Gent DC, Hoeijmakers JH, Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev Genet 2:196–206
Ventura M, Archidiacono N, Rocchi M (2001) Centromere emergence in evolution. Genome Res 11:595–599
Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D’Addabbo P, Wandall A, Bjorck E, de Jong PJ, She X, Eichler EE, Archidiacono N, Rocchi M (2004) Recurrent sites for new centromere seeding. Genome Res 14:1696–1703
Vosman B, Arens P (1997) Molecular characterization of GATA/GACA microsatellite repeats in tomato. Genome 40:25–33
Weide R, Hontelez J, van Kammen A, Koorneef M, Zabel P (1998) Paracentromeric sequences on tomato chromosome 6 show homology to human satellite III and to the mammalian CENP-B binding box. Mol Gen Genet 259:190–197
Witte CP, Le QH, Bureau T, Kumar A (2001) Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A 98:13778–13783
Wong LH, Choo KHA (2001) Centromere on the move. Genome Res 11:513–516
Yang TJ, Yu Y, Nah GJ, Atkins M, Lee S, Frisch DA, Wing RA (2003) Construction and utilities of 10 kb libraries for efficient clone-gap closure for rice genome sequencing. Theor Appl Genet 107:652–660
Yang TJ, Yu Y, Frisch D, Lee S, Kim HR, Kwon SJ, Park BS, Wing RA (2004) Construction of various copy number plasmid vectors and their utility for genome sequencing. Genomics & Informatics 2:153–158
Yang TJ, Yu Y, Lee S, Chang SB, Ahn SN, de Jong JH, Wing RA (2005) Toward finishing rice telomere gap: mapping and sequencing of rice subtelomere regions. Theor Appl Genet (in press)
Yu Y, Rambo T, Currie J, Saski C, Kim HR, Collura K, Thompson S, Simmons J, Yang TJ, Park GN, Patel AJ et al (2003) In-depth view of structure, activity, and evolution of rice chromosome 10 (The Rice Chromosome 10 Sequencing Consortium). Science 300:1566–1569
Zhang HB, Budiman MA, Wing RA (2000) Genetic mapping of jointless-2 to tomato chromosome 12 using RAPD and RFLP analysis. Theor Appl Genet 100:1183–1189
Zhang Y, Huang Y, Zhang L, Li Y, Lu T, Lu Y, Feng Q, Zhao Q, Cheng Z, Xue Y, Wing RA, Han B (2004) Structural features of the rice chromosome 4 centromere. Nucleic Acids Res 32:2023–2030
Zhong XB, de Jong JH, Zabel P (1996) Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res 4:24–28
Zhong XB, Fransz PF, Wennekes-Eden J, Ramanna MS, van Kammen A, Zabel P, de Jong JH (1998) FISH studies reveal the molecular and chromosomal organization of individual telomere domains in tomato. Plant J 13:507–517
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgements
This work was supported by the NSF Plant Genome Grant # 0116076 to R.A.W.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Shaw
The sequence data from this study have been submitted to GenBank under accession no. AY850394
Rights and permissions
About this article
Cite this article
Yang, TJ., Lee, S., Chang, SB. et al. In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114, 103–117 (2005). https://doi.org/10.1007/s00412-005-0342-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-005-0342-8