Abstract
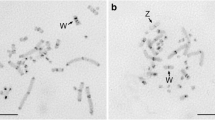
We isolated a new family of satellite DNA sequences from Hae III- and Eco RI-digested genomic DNA of the Blakiston’s fish owl ( Ketupa blakistoni). The repetitive sequences were organized in tandem arrays of the 174 bp element, and localized to the centromeric regions of all macrochromosomes, including the Z and W chromosomes, and microchromosomes. This hybridization pattern was consistent with the distribution of C-band-positive centromeric heterochromatin, and the satellite DNA sequences occupied 10% of the total genome as a major component of centromeric heterochromatin. The sequences were homogenized between macro- and microchromosomes in this species, and therefore intraspecific divergence of the nucleotide sequences was low. The 174 bp element cross-hybridized to the genomic DNA of six other Strigidae species, but not to that of the Tytonidae, suggesting that the satellite DNA sequences are conserved in the same family but fairly divergent between the different families in the Strigiformes. Secondly, the centromeric satellite DNAs were cloned from eight Strigidae species, and the nucleotide sequences of 41 monomer fragments were compared within and between species. Molecular phylogenetic relationships of the nucleotide sequences were highly correlated with both the taxonomy based on morphological traits and the phylogenetic tree constructed by DNA-DNA hybridization. These results suggest that the satellite DNA sequence has evolved by concerted evolution in the Strigidae and that it is a good taxonomic and phylogenetic marker to examine genetic diversity between Strigiformes species.
















Similar content being viewed by others
References
Belterman RHR, de Boer LEM (1984) A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 65:39–82
Chen Z-Q, Lin CC, Hodgetts RB (1989) Cloning and characterization of a tandemly repeated DNA sequence in the crane family (Gruidae). Genome 32:646–654
De Boer LEM (1984) New developments in vertebrate cytotaxonomy VIII. A current list of references on avian karyology. Genetica 65:3–37
Longmire JL, Lewis AK, Brown NC, Buckingham JM, Clark LM, Jones MD, Meincke LJ, Meyne J, Ratliff RL, Ray FA, Wagner RP, Moyzis RK (1988) Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2:14–24
Madsen CS, de Kloet DH, Brooks JE, de Kloet SR (1992) Highly repeated DNA sequences in birds: the structure and evolution of an abundant, tandemly repeated 190-bp DNA fragment in parrots. Genomics 14:462–469
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matzke MA, Varga F, Berger H, Schernthaner J, Schweizer D, Mayr B, Matzke AJM (1990) A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma 99:131–137
Matzke AJM, Varga F, Gruendler P, Unfried I, Berger H, Mayr B, Matzke MA (1992) Characterization of a new repetitive sequence that is enriched on microchromosomes of turkey. Chromosoma 102:9–14
McQueen HA, Fantes J, Cross SH, Clark VH, Archibald AL, Bird AP (1996) CpG islands of chicken are concentrated on microchromosomes. Nat Genet 12:321–324
McQueen HA, Siriaco G, Bird AP (1998) Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res 8:621–630
Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, Ellegren H (1997) Low frequency of microsatellites in the avian genome. Genome Res 7:471-482
Rebholz WER, de Boer LEM, Sasaki M, Belterman RHR, Nishida-Umehara C (1993) The chromosomal phylogeny of owls (Strigiformes) and new karyotypes of seven species. Cytologia 58:403–416
Saifitdinova AF, Derjusheva SE, Malykh AG, Zhurov VG, Andreeva TF, Gaginskaya ER (2001) Centromeric tandem repeat from the chaffinch genome: isolation and molecular characterization. Genome 44:96–103
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasaki M, Takagi N, Nishida C (1984) Current profiles of avian cytogenetics, with notes on chromosomal diagnosis of sex in birds. Nucleus 27:63–73
Sasaki M, Nishida-Umehara C, Tsuchiya K (1994) Interspecific variations in centromeric C-band of the Z chromosome and silver stained nucleolus organizer regions (Ag-NORs) among ten species of owls (Strigiformes). Chrom Inform Serv 56:19–21
Schmutz SM, Moker JS (1991) A cytogenetic comparison of some North American owl species. Genome 34:714-717
Shetty S, Griffin DK, Graves JAM (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7:289–295
Shibusawa M, Nishiboro M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y (2004) Karyotypic evolution in the Galliformes: an examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet Genome Res (in press)
Sibley CG, Ahlquist JE (1990) Phylogeny and classification of birds: a study in molecular evolution. Yale University Press, New Haven
Sibley CG, Monroe Jr BL (1990) Distribution and taxonomy of birds of the world. Yale University Press, New Haven
Singer MF (1982) Highly repeated sequences in mammalian genomes. Int Rev Cytol 76:67–112
Smith J, Bruley CK, Paton IR, Dunn I, Jones CT, Windsor D, Morrice DR, Law AS, Masabanda J, Sazanov A, Waddington D, Fries R, Burt DW (2000) Differences in gene density on chicken macrochromosomes and microchromosomes. Anim Genet 31:96–103
Solovei IV, Joffe BI, Gaginskaya ER, Macgregor HC (1996) Transcription on lampbrush chromosomes of a centromerically localized highly repeated DNA in pigeon ( Columba) relates to sequence arrangement. Chromosome Res 4:588–603
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Suzuki T, Kurosaki T, Agata K, Koide M, Shimada K, Kansaku N, Namikawa T, Matsuda Y (1999a) Cytogenetic assignment of 29 functional genes to chicken microchromosomes by FISH. Cytogenet Cell Genet 87:233-237
Suzuki T, Kurosaki T, Shimada K, Kansaku N, Kuhnlein U, Zadworny D, Agata K, Hashimoto A, Koide M, Koike M, Takata M, Kuroiwa A, Minai S, Namikawa T, Matsuda Y (1999b) Cytogenetic mapping of 31 functional genes on chicken chromosomes by direct R-banding FISH. Cytogenet Cell Genet 87:32–40
Takagi N, Sasaki M (1974) A phylogenetic study of bird karyotypes. Chromosoma 46:91–120
Tanaka K, Suzuki T, Nojiri T, Yamagata T, Namikawa T, Matsuda Y (2000) Characterization and chromosomal distribution of a novel satellite DNA sequence of Japanese quail ( Coturnix coturnix japonica). J Hered 91: 412–415
Yamada K, Nishida-Umehara C, Matsuda Y (2002a) Characterization and chromosomal distribution of novel satellite DNA sequences of the lesser rhea ( Pterocnemia pennata)and the greater rhea ( Rhea americana). Chromosome Res 10:513–523
Yamada K, Shibusawa M, Tsudzuki M, Matsuda Y (2002b) Molecular cloning and characterization of novel centromeric repetitive DNA sequences in the blue-breasted quail ( Coturnix chinensis,Galliformes). Cytogenet Genome Res 98:255–261
Acknowledgements
This study was supported by Grants-in-Aid for Scientific Research (Nos. 11NP0201 and 15370001) from the Ministry of Education, Culture, Sports, Science and Technology in Japan. We used the specimens of Blakiston’s fish owl in this study with the co-operation of the Blakiston’s fish owl conservation programs organized by the Ministry of the Environment, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00412-004-0283-7
Communicated by Y. Hiraoka
Rights and permissions
About this article
Cite this article
Yamada, K., Nishida-Umehara, C. & Matsuda, Y. A new family of satellite DNA sequences as a major component of centromeric heterochromatin in owls (Strigiformes). Chromosoma 112, 277–287 (2004). https://doi.org/10.1007/s00412-003-0267-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-003-0267-z