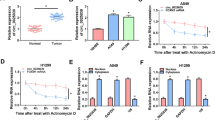
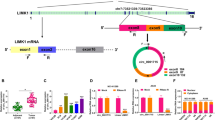
Abstract
Background
Circular RNA migration and invasion inhibitory protein (circMIIP) is reported to be upregulated in non-small cell lung cancer (NSCLC) tissues compared with normal tissues. However, the role and working mechanism of circMIIP in NSCLC progression remain largely unclear.
Methods
Cell proliferation ability was analyzed by colony formation assay, cell counting kit-8 (CCK-8) assay, and 5-ethynyl-2′-deoxyuridine assay. Cell apoptosis was assessed by flow cytometry. Transwell assays were performed to analyze the migration and invasion abilities of NSCLC cells. The interaction between microRNA-766-5p (miR-766-5p) and circMIIP or family with sequence similarity 83A (FAM83A) was validated by dual-luciferase reporter assay and RNA immunoprecipitation assay. Xenograft tumor model was established to analyze the role of circMIIP on tumor growth in vivo.
Results
CircMIIP was highly expressed in NSCLC tissues and cell lines. CircMIIP knockdown restrained the proliferation, migration and invasion and induced the apoptosis of NSCLC cells. CircMIIP acted as a molecular sponge for miR-766-5p, and circMIIP silencing-mediated anti-tumor effects were largely overturned by the knockdown of miR-766-5p in NSCLC cells. miR-766-5p interacted with the 3’ untranslated region (3′UTR) of FAM83A, and FAM83A overexpression largely reversed miR-766-5p accumulation-induced anti-tumor effects in NSCLC cells. CircMIIP competitively bound to miR-766-5p to elevate the expression of FAM83A in NSCLC cells. CircMIIP knockdown significantly restrained xenograft tumor growth in vivo.
Conclusion
CircMIIP promoted cell proliferation, migration and invasion and suppressed cell apoptosis in NSCLC cells through mediating miR-766-5p/FAM83A axis.






Similar content being viewed by others
Data Availability
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Relli V, Trerotola M, Guerra E, Alberti S (2019) Abandoning the notion of non-small cell lung cancer. Trends Mol Med 25(7):585–594
Wang J, Zhou T, Wang T, Wang B (2018) Suppression of lncRNA-ATB prevents amyloid-beta-induced neurotoxicity in PC12 cells via regulating miR-200/ZNF217 axis. Biomed Pharmacother 108:707–715. https://doi.org/10.1016/j.biopha.2018.08.155
Rong G, Wang H, Bowlus CL, Wang C, Lu Y, Zeng Z, Qu J, Lou M, Chen Y, An L, Yang Y, Gershwin ME (2015) Incidence and risk factors for hepatocellular carcinoma in primary biliary cirrhosis. Clin Rev Allergy Immunol 48(2–3):132–141. https://doi.org/10.1007/s12016-015-8483-x
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77(9):2339–2350. https://doi.org/10.1158/0008-5472.can-16-1883
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D (2015) Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 5:8057. https://doi.org/10.1038/srep08057
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY (2017) Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep 7(1):5428. https://doi.org/10.1038/s41598-017-05432-8
Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y, Wang X (2019) The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun 513(4):904–911. https://doi.org/10.1016/j.bbrc.2019.04.108
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG (2018) Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol 144(4):667–674. https://doi.org/10.1007/s00432-017-2576-2
du Manoir S, Guillaud P, Camus E, Seigneurin D, Brugal G (1991) Ki-67 labeling in postmitotic cells defines different Ki-67 pathways within the 2c compartment. Cytometry 12(5):455–463. https://doi.org/10.1002/cyto.990120511
Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H (1984) Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 133(4):1710–1715
Rioux-Leclercq N, Turlin B, Bansard J, Patard J, Manunta A, Moulinoux JP, Guillé F, Ramée MP, Lobel B (2000) Value of immunohistochemical Ki-67 and p53 determinations as predictive factors of outcome in renal cell carcinoma. Urology 55(4):501–505. https://doi.org/10.1016/s0090-4295(99)00550-6
Visapää H, Bui M, Huang Y, Seligson D, Tsai H, Pantuck A, Figlin R, Rao JY, Belldegrun A, Horvath S, Palotie A (2003) Correlation of Ki-67 and gelsolin expression to clinical outcome in renal clear cell carcinoma. Urology 61(4):845–850. https://doi.org/10.1016/s0090-4295(02)02404-4
Huang H (2018) Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors 18:3249. https://doi.org/10.3390/s18103249
Gialeli C, Theocharis AD, Karamanos NK (2011) Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278(1):16–27. https://doi.org/10.1111/j.1742-4658.2010.07919.x
Farina AR, Mackay AR (2014) Gelatinase B/MMP-9 in tumour pathogenesis and progression. Cancers 6(1):240–296. https://doi.org/10.3390/cancers6010240
Xu D, McKee CM, Cao Y, Ding Y, Kessler BM, Muschel RJ (2010) Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Can Res 70(17):6988–6998. https://doi.org/10.1158/0008-5472.can-10-0242
Dong DD, Zhou H, Li G (2015) ADAM15 targets MMP9 activity to promote lung cancer cell invasion. Oncol Rep 34(5):2451–2460. https://doi.org/10.3892/or.2015.4203
Chen Y, Jiang T, Mao A, Xu J (2014) Esophageal cancer stem cells express PLGF to increase cancer invasion through MMP9 activation. Tumour Biol 35(12):12749–12755. https://doi.org/10.1007/s13277-014-2601-x
Jian H, Zhao Y, Liu B, Lu S (2014) SEMA4b inhibits MMP9 to prevent metastasis of non-small cell lung cancer. Tumour Biol 35(11):11051–11056. https://doi.org/10.1007/s13277-014-2409-8
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7(2):e30733. https://doi.org/10.1371/journal.pone.0030733
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S (2017) Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 388(2):08–219. https://doi.org/10.1016/j.canlet.2016.12.006
Feng Y, Wang Q, Shi C, Liu C, Zhang Z (2019) Does circular RNA exert significant effects in ovarian cancer? Crit Rev Eukaryot Gene Expr 29(2):161–170. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2019025941
Chen X, Yu J, Tian H, Shan Z, Liu W, Pan Z, Ren J (2019) Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol 234(11):19130–19140. https://doi.org/10.1002/jcp.28692
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A (2017) Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 23(14):3918–3928. https://doi.org/10.1158/1078-0432.ccr-16-2541
Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I (2018) Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci 75(19):3539–3551. https://doi.org/10.1007/s00018-018-2877-x
Chen Y, Min L, Ren C, Xu X, Yang J, Sun X, Wang T, Wang F, Sun C, Zhang X (2017) miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS ONE 12(2):e0171751. https://doi.org/10.1371/journal.pone.0171751
Jia B, Xia L, Cao F (2018) The role of miR-766-5p in cell migration and invasion in colorectal cancer. Exp Ther Med 15(3):2569–2574. https://doi.org/10.3892/etm.2018.5716
Cai Y, Zhang K, Cao L, Sun H, Wang H (2020) Inhibition of microrna-766-5p attenuates the development of cervical cancer through regulating SCAI. Technol Cancer Res Treat 19:1533033820980081. https://doi.org/10.1177/1533033820980081
Mao Y, Shen G, Su Z, Du J, Xu F, Yu Y (2020) RAD21 inhibited transcription of tumor suppressor MIR4697HG and led to glioma tumorigenesis. Biomed Pharmacother 123:109. https://doi.org/10.1016/j.biopha.2019.109759
Wang M, Liao Q, Zou P (2020) PRKCZ-AS1 promotes the tumorigenesis of lung adenocarcinoma via sponging miR-766-5p to modulate MAPK1. Cancer Biol Ther 21(4):364–371. https://doi.org/10.1080/15384047.2019.1702402
Bai Y, Zhang G, Cheng R, Yang R, Chu H (2019) CASC15 contributes to proliferation and invasion through regulating miR-766-5p/ KLK12 axis in lung cancer. Cell Cycle 18(18):2323–2331. https://doi.org/10.1080/15384101.2019.1646562
Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y, Pang X, Zhang Y, Chen W (2005) BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia 7(12):1073–1080. https://doi.org/10.1593/neo.05406
Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW (2014) Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res 12(8):1156–1165. https://doi.org/10.1158/1541-7786.mcr-13-0289
Bartel CA, Jackson MW (2017) HER2-positive breast cancer cells expressing elevated FAM83A are sensitive to FAM83A loss. PLoS ONE 12(5):e0176778. https://doi.org/10.1371/journal.pone.0176778
Chen S, Huang J, Liu Z, Liang Q, Zhang N, Jin Y (2017) FAM83A is amplified and promotes cancer stem cell-like traits and chemoresistance in pancreatic cancer. Oncogenesis 6(3):e300. https://doi.org/10.1038/oncsis.2017.3
Richtmann S, Wilkens D, Warth A, Lasitschka F, Winter H, Christopoulos P, Herth FJF, Muley T, Meister M, Schneider MA (2019) FAM83A and FAM83B as prognostic biomarkers and potential new therapeutic targets in NSCLC. Cancers. https://doi.org/10.3390/cancers11050652
Zhang J, Sun G, Mei X (2019) Elevated FAM83A expression predicts poorer clincal outcome in lung adenocarcinoma. Cancer Biomark 26(3):367–373. https://doi.org/10.3233/cbm-190520
Zheng YW, Li ZH, Lei L, Liu CC, Wang Z, Fei LR, Yang MQ, Huang WJ, Xu HT (2020) FAM83A promotes lung cancer progression by regulating the Wnt and hippo signaling pathways and indicates poor prognosis. Front Oncol 10:180. https://doi.org/10.3389/fonc.2020.00180
Hu H, Wang F, Wang M, Liu Y, Wu H, Chen X, Lin Q (2020) FAM83A is amplified and promotes tumorigenicity in non-small cell lung cancer via ERK and PI3K/Akt/mTOR pathways. Int J Med Sci 17(6):807–814. https://doi.org/10.7150/ijms.33992
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work. Conceptualization, Methodology, Formal analysis and Data curation: XZ and KW; Validation and Investigation: TW and XZ; Writing—original draft preparation and Writing—review and editing: TW, XZ and KW; Approval of final manuscript: all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The present study was approved by the ethical review committee of Affiliated Hospital of Guizhou Medical University. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
408_2021_500_MOESM2_ESM.tif
Supplementary file2 Sup. Fig 1. CircMIIP is highly expressed in NSCLC tissues and cells. (A) Schematic illustration showing the genomic location of circMIIP generated from its host gene MIIP. (B, C) CircMIIP expression was measured in NSCLC tissues and normal tissues, as well as in NSCLC cell lines (NCI-H23 and A549) and human lung epithelial cell line (BEAS-2B) using RT-qPCR. *P < 0.05 (TIF 3420 kb)
408_2021_500_MOESM3_ESM.tif
Supplementary file3 Sup. Fig 2. CircMIIP silencing restrains the proliferation of NSCLC cells. (A and B) NSCLC cells were transfected with si-NC or si-circMIIP#3. Cell proliferation ability was analyzed by colony formation assay and CCK-8 assay. *P < 0.05 (TIF 3017 kb)
408_2021_500_MOESM4_ESM.tif
Supplementary file4 Sup. Fig 3. The representative images of EdU assay, flow cytometry, and transwell assays in Figure 1. (A) The representative images of EdU assay in Figure 1B. (B) The representative images of flow cytometry in Figure 1C. (C and D) The representative images of transwell assays in Figure 1D and 1E (TIF 22548 kb)
408_2021_500_MOESM5_ESM.tif
Supplementary file5 Sup. Fig 4. CircMIIP absence restrains the proliferation of NSCLC cells largely by up-regulating miR-766-5p. (A and B) NSCLC cells were transfected with si-circMIIP#3 alone or together with anti-miR-766-5p. Colony formation assay and CCK-8 assay were conducted to analyze the proliferation ability of NSCLC cells. *P < 0.05 (TIF 761 kb)
408_2021_500_MOESM6_ESM.tif
Supplementary file6 Sup. Fig 5. miR-766-5p overexpression suppresses the proliferation of NSCLC cells largely by down-regulating FAM83A. (A and B) NSCLC cells were transfected with miR-766-5p alone or together with FAM83A. Cell proliferation ability was analyzed by colony formation assay and CCK-8 assay. *P < 0.05 (TIF 753 kb)
408_2021_500_MOESM7_ESM.tif
Supplementary file7 Sup. Fig 6. CircMIIP/miR-766-5p/FAM83A axis in established in BEAS-2B cells. (A) The knockdown efficiencies of three circMIIP-targeted siRNAs were analyzed in BEAS-2B cells by RT-qPCR. (B) BEAS-2B cells were transfected with si-NC or si-circMIIP#3, and CCK-8 assay was conducted to analyze the proliferation ability of transfected BEAS-2B cells. (C and D) Dual-luciferase reporter assay and RIP assay were conducted to verify the target relationship between circMIIP and miR-766-5p in BEAS-2B cells. (E and F) The interaction between miR-766-5p and FAM83A in BEAS-2B cells was confirmed by dual-luciferase reporter assay and RIP assay. *P < 0.05 (TIF 1432 kb)
408_2021_500_MOESM8_ESM.tif
Supplementary file8 Sup. Fig 7. miR-766-5p overexpression has almost no effect on the proliferation of BEAS-2B cells. (A and B) BEAS-2B cells were transfected with miR-NC or miR-766-5p. (A) RT-qPCR was conducted to analyze the overexpression efficiency of miR-766-5p mimics in BEAS-2B cells. (B) CCK-8 assay was performed to assess the proliferation of BEAS-2B cells. *P < 0.05 (TIF 460 kb)
408_2021_500_MOESM9_ESM.tif
Supplementary file9 Sup. Fig 8. A schematic diagram shows the role of circMIIP/miR-766-5p/FAM83A axis on the malignant behaviors of NSCLC cells (TIF 3065 kb)
Rights and permissions
About this article
Cite this article
Wang, T., Zhu, X. & Wang, K. CircMIIP Contributes to Non-Small Cell Lung Cancer Progression by Binding miR-766-5p to Upregulate FAM83A Expression. Lung 200, 107–117 (2022). https://doi.org/10.1007/s00408-021-00500-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-021-00500-3