Abstract
Purpose
Lots of studies indicated that many microRNAs (miRNAs) are associated with the prognosis of patients with laryngeal cancer (LC). The objective of our study is to identify potential core miRNAs associated with the pathogenesis and prognosis of LC.
Methods
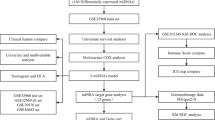
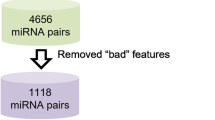
Using the Cancer Genome Atlast database, we identified 70 differentially expressed miRNAs between LC tumor specimens and non-tumor specimens. Then Cox regression analyses and the least absolute shrinkage and selection operator regression signature were performed to detect miRNA prognostic markers. A nomogram integrating miRNA prognostic markers was constructed to predict overall survival (OS) for LC patients. The potential target genes of the key miRNA were predicted by miRTarBase and miRDB databases. Subsequently, their potential functions were revealed by gene ontology annotation and kyoto encyclopedia of genes and genomes pathway enrichment analysis. Related biological pathways of the key target gene involved in LC were detected through gene set enrichment analysis (GSEA).
Results
A prognostic miRNA signature was constructed. The up-regulated miR‐105–1 was related to a worse OS (p = 0.043), which suggested that miR‐105–1 may likely be the key miRNA prognostic marker. Survival analyses and paired expression analyses of target genes indicated that ENDOU may be the key target gene. Finally, we conducted GSEA to elucidate the pathways enriched between low- and high-ENDOU expression datasets.
Conclusion
Our findings might bring some new light on the pathogenesis of LC. Then, it might facilitate doctors to predict the prognosis and improve treatment outcomes for LC patients. However, the behaviors of LC are relatively heterogeneous, and the TCGA database cannot provide detailed information about the subsites and treatment modalities of LC. Further molecular biological experiments and clinical investigations would be required to confirm this conclusion.





Similar content being viewed by others
References
Ho AS, Kim S, Tighiouart M et al (2018) Association of quantitative metastatic lymph node burden with survival in hypopharyngeal and laryngeal cancer. JAMA Oncol 4:985–989
Strieth S, Ernst BP, Both I et al (2019) Randomized controlled single-blinded clinical trial of functional voice outcome after vascular targeting KTP laser microsurgery of early laryngeal cancer. Head Neck 41:899–907
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69:7–34
Shukla GC, Singh J, Barik S (2011) MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 3:83–92
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Fujii T, Shimada K, Nakai T, Ohbayashi C (2018) MicroRNAs in smoking-related carcinogenesis: biomarkers, functions, and therapy. J Clin Med 7:E98
D'Angelo B, Benedetti E, Cimini A, Giordano A (2016) MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res 36:5571–5575
Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20:460–469
Tian L, Cao J, Jiao H et al (2019) CircRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin Sci (Lond) 133:1053–1066
Li P, Yang Y, Liu H et al (2017) MiR-194 functions as a tumor suppressor in laryngeal squamous cell carcinoma by targeting Wee1. J Hematol Oncol 10:32
Garo KS, Miroslavov PT, Stancheva G et al (2020) Novel insights into laryngeal squamous cell carcinoma from association study of aberrantly expressed miRNAs, lncRNAs and clinical features in Bulgarian patients. J BUON 25:357–366
Hutter C, Zenklusen JC (2018) The cancer genome atlas: creating lasting value beyond its data. Cell 173:283–285
Liu J, Lichtenberg T, Hoadley KA et al (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173:400–416
Wang Z, Song Q, Yang Z et al (2019) Construction of immune-related risk signature for renal papillary cell carcinoma. Cancer Med 8:289–304
Berardi G, Morise Z, Sposito C et al (2019) Development of a nomogram to predict outcome after liver resection for hepatocellular carcinoma in child-pugh b cirrhosis. J Hepatol 72:75–84
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550
Gao W, Zhang Y, Niu M et al (2019) Identification of miR-145-5p-centered competing endogenous rna network in laryngeal squamous cell carcinoma. Proteomics 19:e1900020
Zhou ZX, Zhang ZP, Tao ZZ, Tan TZ (2018) MiR-632 promotes laryngeal carcinoma cell proliferation, migration and invasion through negative regulation of GSK3beta. Oncol Res 28:21–31
Cai K, Wang Y, Bao X (2011) MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J Exp Clin Cancer Res 30:73
Gao C, Hu S (2019) miR-506 is a YAP1-dependent tumor suppressor in laryngeal squamous cell carcinoma. Cancer Biol Ther 20:826–836
Gao W, Zhang C, Li W et al (2019) Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther 27:365–379
Karatas OF (2018) Antiproliferative potential of miR-33a in laryngeal cancer Hep-2 cells via targeting PIM1. Head Neck 40:2455–2461
Ottaviani S, Stebbing J, Frampton AE et al (2018) TGF-beta induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat Commun 9:1845
Khalaj M, Woolthuis CM, Hu W et al (2017) miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med 214:2453–2470
Funamizu N, Lacy CR, Kamada M, Yanaga K, Manome Y (2019) microRNA-200b and -301 are associated with gemcitabine response as biomarkers in pancreatic carcinoma cells. Int J Oncol 54:991–1000
Dong P, Xiong Y, Yu J et al (2018) Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene 37:5257–5268
Fang SS, Guo JC, Zhang JH et al (2019) A P53-related microRNA model for predicting the prognosis of hepatocellular carcinoma patients. J Cell Physiol 235:3569–3578
Ma YS, Wu TM, Lv ZW et al (2017) High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget 8:11896–11905
Lu G, Fu D, Jia C et al (2017) Reduced miR-105-1 levels are associated with poor survival of patients with non-small cell lung cancer. Oncol Lett 14:7842–7848
Wei S, Liu K, He Q, Gao Y, Shen L (2019) PES1 is regulated by CD44 in liver cancer stem cells via miR-105-5p. FEBS Lett 593:1777–1786
Zuo JJ, Tao ZZ, Chen C et al (2017) Characteristics of cigarette smoking without alcohol consumption and laryngeal cancer: overall and time-risk relation. A meta-analysis of observational studies. Eur Arch Otorhinolaryngol 274:1617–1631
Kariche N, Hortal MT, Benyahia S et al (2018) Comparative assessment of HPV, alcohol and tobacco etiological fractions in Algerian patients with laryngeal squamous cell carcinoma. Infect Agent Cancer 13:8
Zhu Y, Guo L, Wang S, Yu Q, Lu J (2018) Association of smoking and XPG, CYP1A1, OGG1, ERCC5, ERCC1, MMP2, and MMP9 gene polymorphisms with the early detection and occurrence of laryngeal squamous carcinoma. J Cancer 9:968–977
Cao S, Huang Y, Zhang Q et al (2019) Molecular mechanisms of apoptosis and autophagy elicited by combined treatment with oridonin and cetuximab in laryngeal squamous cell carcinoma. Apoptosis 24:33–45
Lin C, Wang Z, Li L et al (2015) The role of autophagy in the cytotoxicity induced by recombinant human arginase in laryngeal squamous cell carcinoma. Appl Microbiol Biotechnol 99:8487–8494
Teng B, Zhao L, Gao J et al (2017) 20(s)-Protopanaxadiol (PPD) increases the radiotherapy sensitivity of laryngeal carcinoma. Food Funct 8:4469–4477
Dai F, Chen G, Wang Y et al (2019) Identification of candidate biomarkers correlated with the diagnosis and prognosis of cervical cancer via integrated bioinformatics analysis. Onco Targets Ther 12:4517–4532
Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY (2016) Proteomic and metabolic signatures of esophageal squamous cell carcinoma. Curr Cancer Drug Targets. https://doi.org/10.2174/1568009616666160203113721
Biswas NK, Das S, Maitra A, Sarin R, Majumder PP (2014) Somatic mutations in arachidonic acid metabolism pathway genes enhance oral cancer post-treatment disease-free survival. Nat Commun 5:5835
La Paglia L, Listi A, Caruso S et al (2017) Potential role of ANGPTL4 in the cross talk between metabolism and cancer through PPAR signaling pathway. PPAR Res 2017:8187235
Jachimowicz RD, Beleggia F, Isensee J et al (2019) UBQLN4 represses homologous recombination and is overexpressed in aggressive tumors. Cell 176:505–519
Qiu Z, Guo W, Wang Q et al (2015) MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology 149:1587–1598
Acknowledgements
We are grateful to Dr. Hui-Zi Li for his technical support.
Funding
The work was supported by grants from the National Natural Science Foundation of China (No.81172561), the Ministry of Science and Technology of the People’s Republic of China (No. 2014CB541700), and Department of Science and Technology of Zhejiang Province (No. 2015C03035).
Author information
Authors and Affiliations
Contributions
G-J Huang and B-B Y performed the conception and design of this manuscript. G-J Huang and B-B Y performed data analysis and prepared the figure. G-J Huang and B-B Y drafted and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in the work.
Ethical approval
Ethics approval and patient written informed consent were not required because all analyses in our study were mainly performed based on data from the TCGA database.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.





Rights and permissions
About this article
Cite this article
Huang, GJ., Yang, BB. Identification of core miRNA prognostic markers in patients with laryngeal cancer using bioinformatics analysis. Eur Arch Otorhinolaryngol 278, 1613–1626 (2021). https://doi.org/10.1007/s00405-020-06275-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-020-06275-2