Abstract
Purpose
Assortative mating (AM) or preferential mating is known to influence the genetic architecture of the hearing-impaired (HI) population. AM is now seen as a universal phenomenon with individuals seeking partners based on quantitative, qualitative, and behavioral phenotypes. However, the molecular genetic dynamics of AM among the HI tested in real time are limited to the DFNB1 locus.
Methods
A total of 113 HI partners from 82 South Indian families (52 deaf marrying deaf and 30 deaf marrying normal), previously excluded for DFNB1 (GJB2/6) etiology, were screened for SLC26A4 gene (DFNB4) variants.
Results
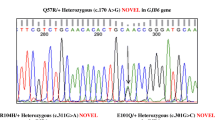
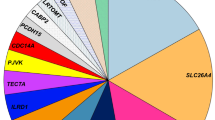
A spectrum of seven pathogenic variants viz., p.S90L, p.V239D, p.V359E, p.Gly389Trpfs*79 (novel), p.T410M, p.N457K and p.K715N were identified. The pathogenic allele frequency of SLC26A4 variants identified in this study was 3.98% (9/226).
Conclusion
We recommend a preliminary screening of mutational hotspots for future investigations to rapidly test for its recurrence among South Indian HI population. This will be the first study to comprehensively account for the incidence of SLC26A4 gene variants and the real-time dynamics of DFNB4 variants among this type of a HI cohort.






Similar content being viewed by others
References
Rendtorff ND, Schrijver I, Lodahl M, Rodriguez-Paris J, Johnsen T, Hansén EC, Tranebjaerg L (2013) SLC26A4 mutation frequency and spectrum in 109 Danish Pendred syndrome/DFNB4 probands and a report of nine novel variants. Clin Genet 84:388–391
Willems PJ (2004) Gene localization and isolation in nonsyndromic hearing loss. In: Willems PJ (ed) Genetic hearing loss. Marcel Dekker Inc., New York, p 203
Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, Riazuddin S (2003) Origins and frequencies of SLC26A4 (PDS) variants in east and south Asians: global implications for the epidemiology of deafness. J Med Genet 40(4):242–248
Zbar RI, Ramesh A, Srisailapathy CS, Fukushima K, Wayne S, Smith RJ (1998) Passage to India: the search for genes causing autosomal recessive nonsyndromic hearing loss. Otolaryngol Head Neck Surg 118(3):333–337
Greinwald JH Jr, Wayne S, Chen AH, Scott DA, Zbar RI, Kraft ML, Lovett M (1998) Localization of a novel gene for nonsyndromic hearing loss (DFNB17) to chromosome region 7q31. Am J Med Genet 78(2):107–113
Van Hauwe P, Everett LA, Coucke P, Scott DA, Kraft ML, Ris-Stalpers C, Ramesh A (1998) Two frequent missense variants in Pendred syndrome. Hum Mol Genet 7(7):1099–1104
Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, Wilcox ER (1998) A mutation in PDS causes non-syndromic recessive deafness. Nat Genet 18(3):215
Adhikary B, Ghosh S, Paul S, Bankura B, Pattanayak AK, Biswas S, Das M (2015) Spectrum and frequency of GJB2, GJB6 and SLC26A4 gene variants among nonsyndromic hearing loss patients in eastern part of India. Gene 573(2):239–245
Pandey N, Rashid T, Jalvi R, Sharma M, Rangasayee R, Andrabi KI, Anand A (2017) Variants in OTOF, CLDN14 & SLC26A4 genes as major causes of hearing impairment in Dhadkai village, Jammu & Kashmir, India. Indian J Med Res 146(4):489
Pavithra A, Jeffrey JM, Chandru J, Ramesh A, Srisailapathy CS (2014) High incidence of GJB2 gene variants among assortatively mating hearing impaired families in Kerala: future implications. J Genet 93(1):207–213
Amritkumar P, Jeffrey JM, Chandru J, Paridhy Vanniya S, Kalaimathi M, Ramakrishnan R, Karthikeyen NP, Srisailapathy CS (2018) Role of DFNB1 variants in hereditary hearing loss among assortative mating hearing impaired families from South India. BMC Med Genet 19(1):105
Morton CC (2002) Genetics, genomics and gene discovery in the auditory system. Hum Mol Genet 11(10):1229–1240
Nance WE, Pandya A (2002) Genetic epidemiology of deafness. In: Genetics of auditory disorders. Springer, New York, pp 67–91
Nance WE, Kearsey MJ (2004) Relevance of connexin deafness (DFNB1) to human evolution. Am J Hum Genet 74(6):1081–1087
Robinson MR, Kleinman A, Graff M, Vinkhuyzen AA, Couper D, Miller MB, van Vliet-Ostaptchouk JV (2017) Genetic evidence of assortative mating in humans. Nat Hum Behav 1(1):0016
Yang T, Gurrola JG II, Wu H, Chiu SM, Wangemann P, Snyder PM, Smith RJ (2009) Variants of KCNJ10 together with variants of SLC26A4 cause digenic nonsyndromic hearing loss associated with enlarged vestibular aqueduct syndrome. Am J Hum Genet 84(5):651–657
Hu H, Wu L, Feng Y, Pan Q, Long Z, Li J, Xia J (2007) Molecular analysis of hearing loss associated with enlarged vestibular aqueduct in the mainland Chinese: a unique SLC26A4 mutation spectrum. J Hum Genet 52(6):492
Zhang F, Xiao Y, Xu L, Zhang X, Zhang G, Li J, Wang H (2016) Mutation analysis of the common deafness genes in patients with nonsyndromic hearing loss in Linyi by SNPscan assay. BioMed Res Int
Jiang H, Chen J, Shan X, Li Y, He J, Yang B (2014) Prevalence and range of GJB2 and SLC26A4 variants in patients with autosomal recessive non-syndromic hearing loss. Mol Med Rep 10:379–386
Zhang M, Han Y, Zhang F, Bai X, Wang H (2019) Mutation spectrum and hotspots of the common deafness genes in 314 patients with nonsyndromic hearing loss in Heze area, China. Acta Oto-Laryngol 139(7):612–617
Koohiyan M (2019) A systematic review of SLC26A4 variants causing hearing loss in the Iranian population. Int J Pediatr Otorhinolaryngol 125:1–5
Vona B, Hofrichter MA, Chioza BA, Crosby AH, Nanda I, Haaf T (2016) Genetic elucidation of nonsyndromic hearing loss in the high-throughput sequencing era. In: Genetics of deafness 2016, vol 20, pp 56–72. Karger Publishers, Basel
Zhao J, Yuan Y, Huang S, Huang B, Cheng J, Kang D, Dai P (2014) KCNJ10 may not be a contributor to nonsyndromic enlargement of vestibular aqueduct (NSEVA) in Chinese subjects. PLoS ONE 9(11):e108134
Blons H, Feldmann D, Duval V, Messaz O, Denoyelle F, Loundon N, Delobel B (2004) Screening of SLC26A4 (PDS) gene in Pendred's syndrome: a large spectrum of variants in France and phenotypic heterogeneity. Clin Genet 66(4):333–340
Bogazzi F, Russo D, Raggi F, Ultimieri F, Berrettini S, Forli F, Bartalena L (2004) Variants in the SLC26A4 (pendrin) gene in patients with sensorineural deafness and enlarged vestibular aqueduct. J Endocrinol Invest 27(5):430–435
Wu CC, Yeh TH, Chen PJ, Hsu CJ (2005) Prevalent SLC26A4 variants in patients with enlarged vestibular aqueduct and/or Mondini dysplasia: a unique spectrum of variants in Taiwan, including a frequent founder mutation. Laryngoscope 115(6):1060–1064
Arellano B, Pera A, Ramirez-Camacho R, Villamar M, Trinidad A, Garcia JR, Hernandez-Chico C (2005) Pendred's syndrome and non-syndromic DFNB4 deafness associated with the homozygous T410M mutation in the SLC26A4 gene in siblings. Clin Genet 67(5):438
Taylor JP, Metcalfe RA, Watson PF, Weetman AP, Trembath RC (2002) Variants of the PDS gene, encoding pendrin, are associated with protein mislocalization and loss of iodide efflux: implications for thyroid dysfunction in Pendred syndrome. J Clin Endocrinol Metab 87(4):1778–1784
Lee HJ, Jung J, Shin JW, Song MH, Kim SH, Lee JH, Lee KY (2014) Correlation between genotype and phenotype in patients with bi-allelic SLC26A4 variants. Clin Genet 86(3):270–275
Kim SY, Kim AR, Kim NK, Lee C, Kim MY, Jeon EH, Park WY, Choi BY (2016) Unraveling of enigmatic hearing-impaired GJB2 single heterozygotes by massive parallel sequencing: DFNB1 or not? Medicine 95(14)
Neverisky DL, Abbott GW (2016) Ion channel–transporter interactions. Crit Rev Biochem Mol Biol 51(4):257–267
Vanniya S P, Chandru J, Pavithra A, Jeffrey JM, Kalaimathi M, Ramakrishnan R, Karthikeyen NP, CR Srikumari S (2018) Recurrence of reported CDH23 variants causing DFNB12 in a special cohort of South Indian hearing impaired assortative mating families—an evaluation. Ann Hum Genet 82(2):119–126
Acknowledgements
We wish to record our profound gratitude to Prof. Alan Bittles, Murdoch University, Australia, for the scientific editing of this manuscript. We are grateful to the hearing-impaired families for their cooperation and participation in this study. This study was supported by major research grants from ad hoc research project (5/8/10-17(Oto)/CFP/2011-NCD-I) from the Indian Council of Medical Research (ICMR) and DBT multicentric grant (BT/PR26850/MED/12/808/2017); UGC Research Scientist Scheme (F.8-17(SC)/90(SA-II)), Government of India, sanctioned to CRS. JMJ was supported by CSIR and currently by DBT SRF; NDG by CSIR SRF. We are thankful to Dr. Suriyakumar Govindarajulu, Anderson Diagnostics & Labs, Chennai, for doing radiological investigations for SLC26A4-positive probands.
Author information
Authors and Affiliations
Contributions
CRS contributed to the conceptualization of this study and obtained funding. CRS, AP, JC, JMJ, and SPV recruited deaf families for the study from four different states, collected detailed clinical data, family history and pedigrees, and isolated DNA from blood samples. AP and JMJ carried out all molecular analysis associated with DFNB1 mutations; JC, SPV, NDG, SM, and JMJ contributed in molecular screening of non-GJB2 gene set. NPK carried out the extended clinical investigations for the SLC26A4-positive families; CRS, JC, and JMJ prepared the original draft and all authors contributed to editing and review and provided intellectual input to the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest between the authors.
Ethics statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional human ethical committee—approval no: UM/IHEC/03-2014-II.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chandru, J., Jeffrey, J.M., Pavithra, A. et al. Genetic analysis of SLC26A4 gene (pendrin) related deafness among a cohort of assortative mating families from southern India. Eur Arch Otorhinolaryngol 277, 3021–3035 (2020). https://doi.org/10.1007/s00405-020-06026-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-020-06026-3