Abstract
Objective
To explore pathogenic/likely pathogenic copy number variations (P/LP CNVs) and regions of homozygosity (ROHs) in fetal central nervous system (CNS) malformations.
Methods
A cohort of 539 fetuses with CNS malformations diagnosed by ultrasound/MRI was retrospectively analyzed between January 2016 and December 2019. All fetuses were analyzed by chromosomal microarray analysis (CMA). Three cases with ROHs detected by CMA were subjected to whole-exome sequencing (WES). The fetuses were divided into two groups according to whether they had other structural abnormalities. The CNS phenotypes of the two groups were further classified as simple (one type) or complicated (≥ 2 types).
Results
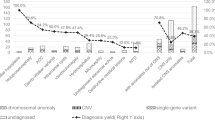
(1) A total of 35 cases with P/LP CNVs were found. The incidence of P/LP CNVs was higher in the extra-CNS group [18.00% (9/50)] than in the isolated group [5.32% (26/489)] (P < 0.01), while there was no significant difference between the simpletype and complicated-type groups. (2) In the simple-type group, the three most common P/LP CNV phenotypes were holoprosencephaly, Dandy–Walker syndrome, and exencephaly. There were no P/LP CNVs associated with anencephaly, microcephaly, arachnoid cysts, ependymal cysts, or intracranial hemorrhage. (3) Only four cases with ROHs were found, and there were no cases of uniparental disomy or autosomal diseases.
Conclusion
The P/LP CNV detection rates varied significantly among the different phenotypes of CNS malformations, although simple CNS abnormalities may also be associated with genetic abnormalities.

Similar content being viewed by others
References
Onkar D, Onkar P, Mitra K (2014) Evaluation of fetal central nervous system anomalies by ultrasound and its anatomical corelation[J]. J Clin Diagn Res 8(6):AC05-07
Rios LT, Araujo Júnior E, Nardozza LM et al (2012) Prenatal and postnatal schizencephaly findings by 2D and 3D ultrasound: pictorial essay[J]. J Clin Imaging Sci 2:30
Bijok J, Dąbkowska S, Kucińska-Chahwan A et al (2022) Prenatal diagnosis of acrania/exencephaly/anencephaly sequence (AEAS): additional structural and genetic anomalies. Arch Gynecol Obstet. https://doi.org/10.1007/s00404-022-06584-3
Goetzinger KR, Stamilio DM, Dicke JM et al (2008) Evaluating the incidence and likelihood ratios for chromosomal abnormalities in fetuses with common central nervous system malformations[J]. Am J Obstet Gynecol 199(3):285.e1–6
Kagan KO, Sonek J, Kozlowski P (2022) Antenatal screening for chromosomal abnormalities. Arch Gynecol Obstet 305(4):825–835
Paul LK, Brown WS, Adolphs R et al (2007) Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity[J]. Nat Rev Neurosci 8(4):287–299
Glass HC, Shaw GM, Ma C et al (2008) Agenesis of the corpus callosum in California 1983–2003: a population-based study[J]. Am J Med Genet A 146A(19):2495–2500
Imataka G, Yamanouchi H, Arisaka O (2007) Dandy-Walker syndrome and chromosomal abnormalities[J]. Congenit Anom (Kyoto) 47(4):113–118
Harper LM, Sutton ALM, Longman RE et al (2014) An economic analysis of prenatal cytogenetic technologies for sonographically detected fetal anomalies. Am J Med Genet Part A 164A(5):1192–1197
American College of Obstetricians and Gynecologists Committee on Genetics (2013) Committee opinion no. 581: the use of chromosomal microarray analysis in prenatal diagnosis[J]. Obstet Gynecol 122(6):1374–1377
Committee on Genetics and the Society for Maternal-Fetal Medicine (2016) Committee opinion no. 682: microarrays and next generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology[J]. Obstet Gynecol 128(6):e262–e268
Yang Y, Muzny DM, Reid JG et al (2013) Clinical whole-exome sequencing for the diagnosis of mendelian disorders[J]. N Engl J Med 369(16):1502–1511
Yang Y, Muzny DM, Xia F et al (2014) Molecular findings among patients referred for clinical whole-exome sequencing[J]. JAMA 312(18):1870–1879
Petrovski S, Aggarwal V, Giordano JL et al (2019) Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study[J]. Lancet 393(10173):758–767
Lord J, McMullan DJ, Eberhardt RY et al (2019) Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study[J]. Lancet 393(10173):747–757
Yang Y, Zhao S, Sun G et al (2022) Genomic architecture of fetal central nervous system anomalies using whole-genome sequencing. NPJ Genom Med 7(1):31
Van den Veyver IB (2019) Prenatally diagnosed developmental abnormalities of the central nervous system and genetic syndromes: a practical review. Prenat Diagn 39(9):666–678
Kui Li, Huixia Y (2022) Comments—Is it appropriate to try intrauterine treatment for fetal hydrocephalus now?[J]. Chin J Perinat Med 25(8):634–635
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels[J]. Bioinformatics 31(16):2745–2747
Kircher M, Witten DM, Jain P et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants[J]. Nat Genet 46(3):310–315
Li L, Fu F, Li R et al (2020) Genetic tests aid in counseling of fetuses with cerebellar vermis defects[J]. Prenat Diagn 40(10):1228–1238
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology[J]. Genet Med 17(5):405–424
Aslan Çetin B, Madazlı R (2022) Assessment of normal fetal cortical sulcus development. Arch Gynecol Obstet 306(3):735–743
Malinger G, Paladini D, Haratz KK et al (2020) ISUOG practice guidelines (updated): sonographic examination of the fetal central nervous system. Part 1: performance of screening examination and indications for targeted neurosonography. Ultrasound Obstet Gynecol 56(3):476–484
Xiaolei X, Fuguang L, Weihe T et al (2019) Correlation between fetal cranial nervous system malformation and chromosome abnormality. Chin J Appl Clin Pediatr 21:1649–1652
Hay SB, Sahoo T, Travis MK, Hovanes K, Dzidic N, Doherty C (2018) Strecker 285 MN. ACOG and SMFM guidelines for prenatal diagnosis: Is karyotyping really 286 sufficient ? [J]. Prenat Diagn 38(3):184–189
Sun L, Wu Q, Jiang SW et al (2015) Prenatal diagnosis of central nervous system anomalies by high-resolution chromosomal microarray analysis[J]. Biomed Res Int 2015:426379
Ishida M, Cullup T, Boustred C et al (2018) A target sequencing panel identifies rare damaging variants in multiple genes in the cranial neural tube defect, anencephaly. Clin Genet 93:870–879
Schumann M, Hofmann A, Krutzke SK et al (2016) Array-based molecular karyotyping in fetuses with isolated brain malformation indentifies disease-causing CNVs. J Neurodev Disord 8:11
Wong K, Moldrich R, Hunter M et al (2015) A familial 7q36.3 duplication associated with agenesis of the corpus callosum[J]. Am J Med Genet A 167A(9):2201–2208
Mimaki M, Shiihara T, Watanabe M et al (2015) Holoprosencephaly with cerebellar vermis hypoplasia in 13q deletion syndrome: critical region for cerebellar dysgenesis within 13q32.2q34[J]. Brain Dev 37(7):714–718
Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound (2012) Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound[J]. Prenat Diagn 32(10):986–995
Santirocco M, Plaja A, Rodó C et al (2021) Chromosomal microarray analysis in fetuses with central nervous system anomalies: an 8-year long observational study from a tertiary care university hospital. Prenat Diagn 41(1):123–135
Song T, Xu Y, Li Y et al (2020) Detection of submicroscopic chromosomal aberrations by chromosomal microarray analysis for the prenatal diagnosis of central nervous system abnormalities. J Clin Lab Anal 34(10):e23434
Xie X, Wu X, Su L et al (2021) Application of single nucleotide polymorphism microarray in prenatal diagnosis of fetuses with central nervous system abnormalities. Int J Gen Med 6(14):4239–4246
Curry CJ, Rosenfeld JA, Grant E et al (2013) The duplication 17p13.3 phenotype: analysis of 21 families delineates developmental, behavioral and brain abnormalities, and rare variant phenotypes[J]. Am J Med Genet A 161A(8):1833–1852
Yamazawa K, Ogata T, Ferguson-Smith AC (2010) Uniparental djsomy and human disease: an overview[J]. Am J Med Genet C Semin Med Genet 154c(3):329–334
Rehder CW, David KL, Hirsch B et al (2013) American college of medical genetics and genomics: standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med 15(2):150–152
Acknowledgements
We thank all the patients who participated in this study.
Funding
This work was supported by the Henan Provincial Health Commission (grant number LHGJ20190361) and by the Key R&D and Promotion Projects in Henan Province (grant number 202102310388).
Author information
Authors and Affiliations
Contributions
YZ, LL and SC: contributed to the conception and design of the study. Material preparation, data collection and analysis were performed by YZ and LL. The critical revision of the manuscript was performed by all authors. All authors commented on the versions of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Approval was granted by the Ethics Committee of the Third Affiliated Hospital of Zhengzhou University (date January 27, 2022/No. 2021-165-01).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhi, Y., Liu, L., Cui, S. et al. Pathogenic/likely pathogenic copy number variations and regions of homozygosity in fetal central nervous system malformations. Arch Gynecol Obstet 308, 1723–1735 (2023). https://doi.org/10.1007/s00404-022-06866-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-022-06866-w