Abstract
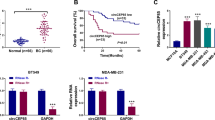
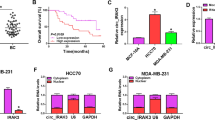
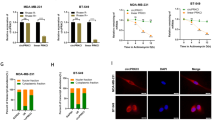
Growing evidence has revealed that circular RNAs (circRNA) play critical roles in cancer progression. Here, we examined the function of a novel circRNA, Circ_0005795, in basal cell carcinoma (BCC) and explored the possible molecular mechanism. Nodular BCC and adjacent non-tumor tissues derived from 30 patients and 2 BCC cell lines were applied to analyze gene expression. Circ_0005795 loss- and gain-of-function were constructed to investigate BCC progression. Nuclear and cytoplasmic fractionation and luciferase assay were carried out to determine cellular localization and molecular interaction of Circ_0005795. Circ_0005795 expression was significantly elevated in BCC tissues and cells. Knockdown of Circ_0005795 dramatically reduced cell viability, colony formation, and anti-apoptotic protein levels, while increased caspase-3 activity. Circ_0005795 located in cytoplasm, which exerted its tumor-promoting effect through targeting and sponging miR-1231 in BCC cells. In summary, Circ_0005795 works as an oncogene in BCC, which might be used as a promising biomarker and a potential therapeutic target for BCC diagnosis and treatment.





Similar content being viewed by others
References
Cameron MC, Lee E, Hibler BP, Barker CA, Mori S, Cordova M, Nehal KS, Rossi AM (2019) Basal cell carcinoma: epidemiology; pathophysiology; clinical and histological subtypes; and disease associations. J Am Acad Dermatol 80:303–317. https://doi.org/10.1016/j.jaad.2018.03.060
Chen G, Shi Y, Liu M, Sun J (2018) circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 9:175. https://doi.org/10.1038/s41419-017-0204-3
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211. https://doi.org/10.1038/nrm.2015.32
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Chen T, Yang YJ, Li YK, Liu J, Wu PF, Wang F, Chen JG, Long LH (2016) Chronic administration tetrahydroxystilbene glucoside promotes hippocampal memory and synaptic plasticity and activates ERKs, CaMKII and SIRT1/miR-134 in vivo. J Ethnopharmacol 190:74–82. https://doi.org/10.1016/j.jep.2016.06.012
Ebbesen KK, Hansen TB, Kjems J (2017) Insights into circular RNA biology. RNA Biol 14:1035–1045. https://doi.org/10.1080/15476286.2016.1271524
Ebert MS, Sharp PA (2010) MicroRNA sponges: progress and possibilities. RNA 16:2043–2050. https://doi.org/10.1261/rna.2414110
Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66:1151–1164. https://doi.org/10.1002/hep.29270
Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J (2018) Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun 503:863–869. https://doi.org/10.1016/j.bbrc.2018.06.088
Khafaei M, Rezaie E, Mohammadi A, Shahnazi Gerdehsang P, Ghavidel S, Kadkhoda S, Zorrieh Zahra A, Forouzanfar N, Arabameri H, Tavallaie M (2019) miR-9: from function to therapeutic potential in cancer. J Cell Physiol. https://doi.org/10.1002/jcp.28210
Kiiski V, de Vries E, Flohil SC, Bijl MJ, Hofman A, Stricker BH, Nijsten T (2010) Risk factors for single and multiple basal cell carcinomas. Arch Dermatol 146:848–855. https://doi.org/10.1001/archdermatol.2010.155
Kleaveland B, Shi CY, Stefano J, Bartel DP (2018) A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174(350–362):e317. https://doi.org/10.1016/j.cell.2018.05.022
Li W, Li Y, Sun Z, Zhou J, Cao Y, Ma W, Xie K, Yan X (2019) Comprehensive circular RNA profiling reveals the regulatory role of the hsa_circ_0137606/miR1231 pathway in bladder cancer progression. Int J Mol Med 44:1719–1728. https://doi.org/10.3892/ijmm.2019.4340
Li X, Yang L, Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442. https://doi.org/10.1016/j.molcel.2018.06.034
Luan W, Shi Y, Zhou Z, Xia Y, Wang J (2018) circRNA_0084043 promote malignant melanoma progression via miR-153-3p/snail axis. Biochem Biophys Res Commun 502:22–29. https://doi.org/10.1016/j.bbrc.2018.05.114
Madan V, Hoban P, Strange RC, Fryer AA, Lear JT (2006) Genetics and risk factors for basal cell carcinoma. Br J Dermatol 154(Suppl 1):5–7. https://doi.org/10.1111/j.1365-2133.2006.07229.x
Mattick JS (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. BioEssays 25:930–939. https://doi.org/10.1002/bies.10332
Nagarajan P, Asgari MM, Green AC, Guhan SM, Arron ST, Proby CM, Rollison DE, Harwood CA, Toland AE (2019) Keratinocyte carcinomas: current concepts and future research priorities. Clin Cancer Res 25:2379–2391. https://doi.org/10.1158/1078-0432.CCR-18-1122
Nehal KS, Bichakjian CK (2018) Update on keratinocyte carcinomas. N Engl J Med 379:363–374. https://doi.org/10.1056/NEJMra1708701
Otto T, Sicinski P (2017) Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17:93–115. https://doi.org/10.1038/nrc.2016.138
Quan Z, Zhang BB, Yin F, Du J, Zhi YT, Xu J, Song N (2019) DDX5 silencing suppresses the migration of basal cell carcinoma cells by downregulating JAK2/STAT3 pathway. Technol Cancer Res Treat 18:1533033819892258. https://doi.org/10.1177/1533033819892258
Que SKT, Zwald FO, Schmults CD (2018) Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 78:237–247. https://doi.org/10.1016/j.jaad.2017.08.059
Shi YR, Wu Z, Xiong K, Liao QJ, Ye X, Yang P, Zu XB (2020) Circular RNA circKIF4A sponges miR-375/1231 to promote bladder cancer progression by upregulating NOTCH2 expression. Front Pharmacol 11:605. https://doi.org/10.3389/fphar.2020.00605
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, Kjems J, Laybutt DR, Regazzi R (2018) Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab 9:69–83. https://doi.org/10.1016/j.molmet.2018.01.010
Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, Liu Y, Liu L, Wei YQ, Peng Y (2018) Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res 28:693–695. https://doi.org/10.1038/s41422-018-0033-7
Totonchy M, Leffell D (2017) Emerging concepts and recent advances in basal cell carcinoma. F1000Res 6:2085. https://doi.org/10.12688/f1000research.11314.1
Tsujimoto Y (1998) Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria? Genes Cells 3:697–707. https://doi.org/10.1046/j.1365-2443.1998.00223.x
Verkouteren JAC, Ramdas KHR, Wakkee M, Nijsten T (2017) Epidemiology of basal cell carcinoma: scholarly review. Br J Dermatol 177:359–372. https://doi.org/10.1111/bjd.15321
Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H (2018) CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res 78:4812–4825. https://doi.org/10.1158/0008-5472.CAN-18-0532
Wu S, Han J, Li WQ, Li T, Qureshi AA (2013) Basal-cell carcinoma incidence and associated risk factors in U.S. women and men. Am J Epidemiol 178:890–897. https://doi.org/10.1093/aje/kwt073
Yu CY, Kuo HC (2019) The emerging roles and functions of circular RNAs and their generation. J Biomed Sci 26:29. https://doi.org/10.1186/s12929-019-0523-z
Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L (2016) The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE 11:e0158347. https://doi.org/10.1371/journal.pone.0158347
Zhang J, Zhang J, Qiu W, Zhang J, Li Y, Kong E, Lu A, Xu J, Lu X (2018) MicroRNA-1231 exerts a tumor suppressor role through regulating the EGFR/PI3K/AKT axis in glioma. J Neurooncol 139:547–562. https://doi.org/10.1007/s11060-018-2903-8
Zhao ZJ, Shen J (2017) Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 14:514–521. https://doi.org/10.1080/15476286.2015.1122162
Zhong Z, Lv M, Chen J (2016) Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 6:30919. https://doi.org/10.1038/srep30919
Zhu H, Wu J, Cui X, Chen X (2017) Bmi-1 serves as a potential novel marker for progression in human cutaneous basal cell carcinoma. Int J Clin Exp Pathol 10:8928–8935
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All the patients were obtained informed consent following a protocol approved by the ethics board of Qingdao Municipal Hospital, and this study was conducted in accordance with the ethical standards formulated in the Declaration of Helsinki.
Informed consent
All participants in this study were informed and gave a written consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Y., Li, Y. & Li, L. Circular RNA hsa_Circ_0005795 mediates cell proliferation of cutaneous basal cell carcinoma via sponging miR-1231. Arch Dermatol Res 313, 773–782 (2021). https://doi.org/10.1007/s00403-020-02174-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-020-02174-y