Abstract
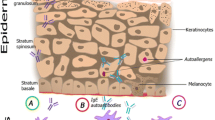
Atopic dermatitis (AD) is a skin disease that results from a combination of skin barrier dysfunction and immune dysregulation. The immune dysregulation is often associated with IgE sensitivity. There is also evidence that autoallergens Hom s 1, 2, 3, and 4 play a role in AD; it is possible that patients with specific HLA subtypes are predisposed to autoreactivity due to increased presentation of autoallergen peptides. The goal of our study was to use in silico epitope prediction platforms as an approach to identify HLA subtypes that may preferentially bind autoallergen peptides and are thus candidates for further study. Considering the previously described association of DRB1 alleles with AD and progression of disease, emphasis was placed on DRB1. Certain DRB1 alleles (08:04, 11:01, and 11:04) were identified by both algorithms to bind a significant percent of the generated autoallergen peptides. Conversely, autoallergen core peptide sequences FRQLSHRFH and IRAKLRLQA (Hom s 1), IRKSKNILF (Hom s 2), FKWVPVTDS and MAAIEKVRK (Hom s 3), and FRYFATLKV (Hom s 4) were predicted to bind many DRB1 alleles and, thus, may play a role in the pathogenesis of AD. Our findings provide candidate DRB1 alleles and autoallergen epitopes that will guide future studies exploring the relationship between DRB1 subtype and autoreactivity in AD. A similar approach can be used for any antigen that has been associated with an IgE response and AD.

Similar content being viewed by others
Abbreviations
- AD:
-
Atopic dermatitis
- IEDB:
-
Immune Epitope Database Analysis Resource
- HA:
-
Hemagglutinin
References
Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S (2010) NetMHCIIpan-2.0—improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res 6:9
Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S et al (2008) Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol 4(7):e1000107
Paul S, Kolla RV, Sidney J, Weiskopf D, Fleri W, Kim Y, et al (2013) Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clin Dev Immunol, pp 1–7 (ID 467852)
Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE et al (2008) Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res 36(Web Server issue):513–518
Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M (2013) NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 65(10):711–724
Nielsen M, Lund O (2009) NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform 10:296
Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH et al (2010) Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol 185(2):943–955
Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH et al (2012) Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production. J Immunol 189(2):679–688
Ramesh M, Yuenyongviwat A, Konstantinou GN, Lieberman J, Pascal M, Masilamani M et al (2016) Peanut T-cell epitope discovery: Ara h 1. J Allergy Clin Immunol 137(6):1764–71.e4
Schulten V, Greenbaum JA, Hauser M, McKinney DM, Sidney J, Kolla R et al (2013) Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci USA 110(9):3459–3464
Tangri S, Mothe BR, Eisenbraun J, Sidney J, Southwood S, Briggs K et al (2005) Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol 174(6):3187–3196
Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M et al (2010) Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinform 11:568
Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S et al (1998) Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 160(7):3363–3373
Valenta R (2002) The future of antigen-specific immunotherapy of allergy. Nat Rev Immunol 2(6):446–453
Wills-Karp M, Santeliz J, Karp CL (2001) The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol 1(1):69–75
Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P et al (2006) Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy Asthma and Immunology/PRACTALL Consensus Report. Allergy 61(8):969–987
Leung DY, Bieber T (2003) Atopic dermatitis. [Review] [100 refs]. Lancet 361(9352):151–160
Hay RJ, Johns NE, Williams HC, Bolliger IW, Dellavalle RP, Margolis DJ et al (2014) The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Investig Dermatol 134(6):1527–1534
Margolis JS, Abuabrar K, Bilker W, Hoffstad O, Margolis DJ (2014) Persistance of mild of mild to moderate atopic dermatitis. JAMA Dermatol 150(6):593–600
Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al (2013) Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. (Erratum appears in Lancet. 2013;381(9867):628 Note: AlMazroa, Mohammad A [added]; Memish, Ziad A [added]]. Lancet. 2012;380(9859):2197–223)
Leung DY (2000) Atopic dermatitis: new insights and opportunities for therapeutic intervention. J Allergy Clin Immunol 105(5):860–876
Leung DY (2013) New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol Int 62(2):151–161
Eyerich K, Novak N (2013) Immunology of atopic eczema: overcoming the Th1/Th2 paradigm. Allergy 68(8):974–982
Eyerich S, Onken AT, Weidinger S, Franke A, Nasorri F, Pennino D et al (2011) Mutual antagonism of T cells causing psoriasis and atopic eczema. N Engl J Med 365(3):231–238
Gittler JK, Shemer A, Suarez-Farinas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQF et al (2012) Progressive activation of Th2/Th22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol 130:1344–1354
Brandt EB, Sivaprasad U (2011) Th2 cytokines and atopic dermatitis. J Clin Cell Immunol 2(3):110
Grewe M, Walther S, Gyufko K, Czech W, Schopf E, Krutmann J (1995) Analysis of the cytokine pattern expressed in situ in inhalant allergen patch test reactions of atopic dermatitis patients. J Invest Dermatol 105(3):407–410
Howell MD, Fairchild HR, Kim BE, Bin L, Boguniewicz M, Redzic JS et al (2008) Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J Invest Dermatol 128(9):2248–2258
Jurgens M, Wollenberg A, Hanau D, de la Salle H, Bieber T (1995) Activation of human epidermal Langerhans cells by engagement of the high affinity receptor for IgE, Fc epsilon RI. J Immunol 155(11):5184–5189
Maurer D, Ebner C, Reininger B, Fiebiger E, Kraft D, Kinet JP et al (1995) The high affinity IgE receptor (Fc epsilon RI) mediates IgE-dependent allergen presentation. J Immunol 154(12):6285–6290
Valenta R, Maurer D, Steiner R, Seiberler S, Sperr WR, Valent P et al (1996) Immunoglobulin E response to human proteins in atopic patients. J Invest Dermatol 107(2):203–208
Valenta R, Seiberler S, Natter S, Mahler V, Mossabeb R, Ring J et al (2000) Autoallergy: a pathogenetic factor in atopic dermatitis? J Allergy Clin Immunol 105(3):432–437
Natter S, Seiberler S, Hufnagl P, Binder BR, Hirschl AM, Ring J et al (1998) Isolation of cDNA clones coding for IgE autoantigens with serum IgE from atopic dermatitis patients. Faseb J 12(14):1559–1569
Valenta R, Natter S, Seiberler S, Wichlas S, Maurer D, Hess M et al (1998) Molecular characterization of an autoallergen, Hom s 1, identified by serum IgE from atopic dermatitis patients. J Invest Dermatol 111(6):1178–1183
Tang TS, Bieber T, Williams HC (2012) Does "autoreactivity" play a role in atopic dermatitis? J Allergy Clin Immunol 129(5):1209–15.e2
Roesner LM, Werfel T (2019) Autoimmunity (or not) in atopic dermatitis. Front Immunol 10:2128
Aichberger KJ, Mittermann I, Reininger R, Seiberler S, Swoboda I, Spitzauer S et al (2005) Hom s 4, an IgE-reactive autoantigen belonging to a new subfamily of calcium-binding proteins, can induce Th cell type 1-mediated autoreactivity. J Immunol 175(2):1286–1294
Nielsen M, Lundegaard C, Lund O (2007) Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform 8:238
Roesner LM, Heratizadeh A, Wieschowski S, Mittermann I, Valenta R, Eiz-Vesper B et al (2016) Alpha-NAC-specific autoreactive CD8+ T cells in atopic dermatitis are of an effector memory type and secrete IL-4 and IFN-gamma. J Immunol 196(8):3245–3252
Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA et al (2005) Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics 57(5):304–314
Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U et al (1999) Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol 17(6):555–561
Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4(4):e1000048
Maiers M, Gragert L, Klitz W (2007) High resolution HLA alleles and haplotypes in the US population. Hum Immunol 68:779–788
Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT et al (2010) Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci USA 107(28):12599–12604
Jahn-Schmid B, Kelemen P, Himly M, Bohle B, Fischer G, Ferreira F et al (2002) The T cell response to Art v 1, the major mugwort pollen allergen, is dominated by one epitope. J Immunol 169(10):6005–6011
Van Hemelen D, Mahler V, Fischer G, Fae I, Reichl-Leb V, Pickl W et al (2015) HLA class II peptide tetramers vs allergen-induced proliferation for identification of allergen-specific CD4 T cells. Allergy 70(1):49–58
Duvvuri VR, Duvvuri B, Jamnik V, Gubbay JB, Wu J, Wu GE (2013) T cell memory to evolutionarily conserved and shared hemagglutinin epitopes of H1N1 viruses: a pilot scale study. BMC Infect Dis 13:204
Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C et al (2013) Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccin Immunother 9(7):1598–1607
Yang J, James E, Gates TJ, DeLong JH, LaFond RE, Malhotra U et al (2013) CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol 25(8):447–457
Godkin A, Davenport M, Hill AV (2005) Molecular analysis of HLA class II associations with hepatitis B virus clearance and vaccine nonresponsiveness. Hepatology 41(6):1383–1390
Thio CL, Thomas DL, Karacki P, Gao X, Marti D, Kaslow RA et al (2003) Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol 77(22):12083–12087
Menzies R, Vissandjee B, Rocher I, St GY (1994) The booster effect in two-step tuberculin testing among young adults in Montreal. Ann Intern Med 120(3):190–198
Selvaraj P, Reetha AM, Uma H, Xavier T, Janardhanam B, Prabhakar R et al (1996) Influence of HLA-DR and -DO phenotypes on tuberculin reactive status in pulmonary tuberculosis patients. Tuber Lung Dis 77(4):369–373
Brown GR, Hem V, Katz KS, Ovetsky M, Wallin C, Ermolaeva O et al (2015) Gene: a gene-centered information resource at NCBI. Nucleic Acids Res 43(1):D36–42
Androulakis IP, Nayak NN, Ierapetritou MG, Mono DS, Floudas CA (1997) A predictive method for the evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins Struct Funct Genet 29:87–102
Margolis DJ, Mitra N, Kim B, Gupta J, Hoffstad OJ, Papadopoulos M et al (2015) Association of HLA-DRB1 genetic variants with the persistence of atopic dermatitis. Hum Immunol 76(8):571–577
Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG (2015) The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43(Database issue):D423–431
Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodriguez E, Matanovic A, Marenholz I et al (2013) High-density genotyping study indentifies four new susceptibility loci for atopic dermatitis. Nat Genet 45:808–812
Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G et al (2009) A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet 41(5):596–601
Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M et al (2012) Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet 44(11):1222–1226
Mansur AH, Williams GA, Bishop DT, Markham AF, Lewis S, Britton J et al (2000) Evidence for a role of HLA DRB1 alleles in the control of IgE levels, strengthened by interacting TCR A/D marker alleles. Clin Exp Allergy 30(10):1371–1378
Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP et al (2015) Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet 47(12):1449–1456
Potaczek DP, Kabesxh M (2012) Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy 42:852–871
Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF et al (2011) Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet 43(7):690–694
Torres-Galvan MJ, Quiralte J, Blanco C, Castillo R, Carrillo T, Perez-Aciego P et al (2000) Pocket 4 in the HLA-DRB1 antigen-binding groove: an association with atopy. Allergy 55(4):398–401
Weidinger S, Willis-Owen SA, Kamatani Y, Baurecht H, Morar N, Liang L et al (2013) A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum Mol Genet 22(23):4841–4856
Leung CS (2015) Endogenous antigen presentation of MHC class II epitopes through non-autophagic pathways. Front Immunol 6:464
Park H, Ahn K, Park MH, Lee SI (2012) The HLA-DRB1 polymorphism is associated with atopic dermatitis, but not egg allergy in Korean children. Allergy Asthma Immunol Res 4(3):143–149
Altschul SF, Koonin EV (1998) Iterated profile searches with PSI-BLAST–a tool for discovery in protein databases. Trends Biochem Sci 23(11):444–447
Acknowledgements
We thank Nandita Mitra and Ole Hoffstad for their helpful input. This work was supported by the University of Pennsylvania Center for Clinical Epidemiology and Biostatistics Summer Research Fellowship.
Author information
Authors and Affiliations
Contributions
JG, DM, and DJM designed the research study. JG performed the research and wrote the initial draft manuscript. All authors revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gong, J.J., Margolis, D.J. & Monos, D.S. Predictive in silico binding algorithms reveal HLA specificities and autoallergen peptides associated with atopic dermatitis. Arch Dermatol Res 312, 647–656 (2020). https://doi.org/10.1007/s00403-020-02059-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-020-02059-0