Abstract
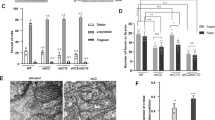
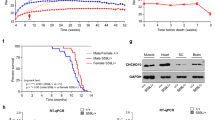
Mutations in coiled-coil-helix–coiled-coil-helix domain containing 10 (CHCHD10), a mitochondrial protein of unknown function, cause a disease spectrum with clinical features of motor neuron disease, dementia, myopathy and cardiomyopathy. To investigate the pathogenic mechanisms of CHCHD10, we generated mutant knock-in mice harboring the mouse-equivalent of a disease-associated human S59L mutation, S55L in the endogenous mouse gene. CHCHD10S55L mice develop progressive motor deficits, myopathy, cardiomyopathy and accelerated mortality. Critically, CHCHD10 accumulates in aggregates with its paralog CHCHD2 specifically in affected tissues of CHCHD10S55L mice, leading to aberrant organelle morphology and function. Aggregates induce a potent mitochondrial integrated stress response (mtISR) through mTORC1 activation, with elevation of stress-induced transcription factors, secretion of myokines, upregulated serine and one-carbon metabolism, and downregulation of respiratory chain enzymes. Conversely, CHCHD10 ablation does not induce disease pathology or activate the mtISR, indicating that CHCHD10S55L-dependent disease pathology is not caused by loss-of-function. Overall, CHCHD10S55L mice recapitulate crucial aspects of human disease and reveal a novel toxic gain-of-function mechanism through maladaptive mtISR and metabolic dysregulation.










Similar content being viewed by others
References
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2014) A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 137:2329–2345. https://doi.org/10.1093/brain/awu138
BonDurant LD, Ameka M, Naber MC, Markan KR, Idiga SO, Acevedo MR et al (2017) FGF21 regulates metabolism through adipose-dependent and -independent mechanisms. Cell Metab 25(935–944):e934. https://doi.org/10.1016/j.cmet.2017.03.005
Brockmann SJ, Freischmidt A, Oeckl P, Muller K, Ponna SK, Helferich AM et al (2018) CHCHD10 mutations p. R15L and p.G66 V cause motoneuron disease by haploinsufficiency. Hum Mol Genet 27:706–715. https://doi.org/10.1093/hmg/ddx436
Burstein SR, Valsecchi F, Kawamata H, Bourens M, Zeng R, Zuberi A et al (2018) In vitro and in vivo studies of the ALS-FTLD protein CHCHD10 reveal novel mitochondrial topology and protein interactions. Hum Mol Genet 27:160–177. https://doi.org/10.1093/hmg/ddx397
Cavallaro G (2010) Genome-wide analysis of eukaryotic twin CX9C proteins. Mol BioSyst 6:2459–2470. https://doi.org/10.1039/c0mb00058b
Crawley JN (1999) Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res 835:18–26
Custer SK, Neumann M, Lu H, Wright AC, Taylor JP (2010) Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum Mol Genet 19:1741–1755. https://doi.org/10.1093/hmg/ddq050ddq050
DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348:2656–2668
Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A et al (2004) Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol 185:232–240
Fleming SM, Salcedo J, Fernagut PO, Rockenstein E, Masliah E, Levine MS et al (2004) Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J Neurosci 24:9434–9440. https://doi.org/10.1523/JNEUROSCI.3080-04.2004
Fratter CDE, Carver J, Sergeant K, Barbosa IA, Hofer M, Esiri M et al (2017) Mitochondrial disease and lipid storage myopathy due to mutation in CHCHD10 or DNM1L and disordered mitochondrial dynamics. Neuromusc Disord 27S1:S21
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Genin EC, Plutino M, Bannwarth S, Villa E, Cisneros-Barroso E, Roy M et al (2016) CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med 8:58–72. https://doi.org/10.15252/emmm.201505496
Gostimskaya I, Galkin A (2010) Preparation of highly coupled rat heart mitochondria. J Vis Exp. https://doi.org/10.3791/2202
Huang X, Wu BP, Nguyen D, Liu YT, Marani M, Hench J et al (2018) CHCHD2 accumulates in distressed mitochondria and facilitates oligomerization of CHCHD10. Hum Mol Genet. https://doi.org/10.1093/hmg/ddy270
Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A et al (2013) mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342:1524–1528. https://doi.org/10.1126/science.1244360
Kawamata H, Manfredi G (2017) Proteinopathies and OXPHOS dysfunction in neurodegenerative diseases. J Cell Biol 216:3917–3929. https://doi.org/10.1083/jcb.201709172
Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S et al (2017) mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab 26(419–428):e415. https://doi.org/10.1016/j.cmet.2017.07.007
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. https://doi.org/10.1186/gb-2013-14-4-r36
Leger M, Quiedeville A, Bouet V, Haelewyn B, Boulouard M, Schumann-Bard P et al (2013) Object recognition test in mice. Nat Protoc 8:2531–2537. https://doi.org/10.1038/nprot.2013.155
Lehmer C, Schludi MH, Ransom L, Greiling J, Junghanel M, Exner N et al (2018) A novel CHCHD10 mutation implicates a Mia40-dependent mitochondrial import deficit in ALS. EMBO Mol Med. https://doi.org/10.15252/emmm.201708558
Li YR, King OD, Shorter J, Gitler AD (2013) Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201:361–372. https://doi.org/10.1083/jcb.201302044
Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438. https://doi.org/10.1016/j.neuron.2013.07.033
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Martineau E, Di Polo A, Vande Velde C, Robitaille R (2018) Dynamic neuromuscular remodeling precedes motor-unit loss in a mouse model of ALS. Elife. https://doi.org/10.7554/elife.41973
Melber A, Haynes CM (2018) UPR(mt) regulation and output: a stress response mediated by mitochondrial–nuclear communication. Cell Res 28:281–295. https://doi.org/10.1038/cr.2018.16
Milner TA, Waters EM, Robinson DC, Pierce JP (2011) Degenerating processes identified by electron microscopic immunocytochemical methods. Methods Mol Biol 793:23–59. https://doi.org/10.1007/978-1-61779-328-8_3
Palomo GM, Granatiero V, Kawamata H, Konrad C, Kim M, Arreguin AJ et al (2018) Parkin is a disease modifier in the mutant SOD1 mouse model of ALS. EMBO Mol Med. https://doi.org/10.15252/emmm.201808888
Perrone F, Nguyen HP, Van Mossevelde S, Moisse M, Sieben A, Santens P et al (2017) Investigating the role of ALS genes CHCHD10 and TUBA4A in Belgian FTD-ALS spectrum patients. Neurobiol Aging 51:177.e9–177.e16. https://doi.org/10.1016/j.neurobiolaging.2016.12.008
Purandare N, Somayajulu M, Huttemann M, Grossman LI, Aras S (2018) The cellular stress proteins CHCHD10 and MNRR1 (CHCHD2): partners in mitochondrial and nuclear function and dysfunction. J Biol Chem 293:6517–6529. https://doi.org/10.1074/jbc.RA117.001073
Purice MD, Taylor JP (2018) Linking hnRNP function to ALS and FTD pathology. Front Neurosci 12:326. https://doi.org/10.3389/fnins.2018.00326
Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D et al (2017) Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 216:2027–2045. https://doi.org/10.1083/jcb.201702058
Sfakianos AP, Mellor LE, Pang YF, Kritsiligkou P, Needs H, Abou-Hamdan H et al (2018) The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ 25:1766–1780. https://doi.org/10.1038/s41418-018-0076-9
Siegmund SE, Yang H, Sharma R, Javors M, Skinner O, Mootha V et al (2017) Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Hum Mol Genet 26:4588–4605. https://doi.org/10.1093/hmg/ddx341
Straub IR, Janer A, Weraarpachai W, Zinman L, Robertson J, Rogaeva E et al (2018) Loss of CHCHD10-CHCHD2 complexes required for respiration underlies the pathogenicity of a CHCHD10 mutation in ALS. Hum Mol Genet 27:178–189. https://doi.org/10.1093/hmg/ddx393
Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M et al (2011) FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol 10:806–818. https://doi.org/10.1016/S1474-4422(11)70155-7
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. https://doi.org/10.1038/nbt.2450
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Woo JA, Liu T, Trotter C, Fang CC, De Narvaez E, LePochat P et al (2017) Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat Commun 8:15558. https://doi.org/10.1038/ncomms15558
Acknowledgements
We acknowledge the funding support of Muscular Dystrophy Association Grant MDA382033 (to G. M.) for this project. We also acknowledge The Jackson Laboratory Genome Engineering Technology and Physiology cores. Costs were defrayed by Cancer Center Support, National Cancer Institute (Grant CA034196) to The Jackson Laboratory. Additional studies were supported using Grant NIH Precision Genetics U54 OD020351 (to C. L.) and NIH/NINDS R01NS062055 (to G. M.). We also acknowledge the WCMC’s Center of Comparative Medicine and Pathology, the Neuroanatomy EM Core in the BMRI, and the EM Imaging Core of WCM.
Author information
Authors and Affiliations
Contributions
GM and CL conceived the study. GM, CL, CJA, AP, TAM, HK, and SRB contributed to experimental design. CL, AZ, CD, and LC designed and performed gene editing and mouse phenotyping. CJA, KB, and SRB performed behavioral experiments. CD and LC performed echocardiography experiments. CJA, TAM, AP, and KB performed histology and electron microscopy experiments. CJA, KB, HK, JD, and SM performed immunohistochemistry and biochemical experiments. GM and CJA analyzed data and drafted the manuscript with input from other authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 3 (MP4 14128 kb)
Supplementary material 4 (MP4 18681 kb)
Supplementary material 5 (MP4 2830 kb)
Supplementary material 6 (MP4 4635 kb)
Rights and permissions
About this article
Cite this article
Anderson, C.J., Bredvik, K., Burstein, S.R. et al. ALS/FTD mutant CHCHD10 mice reveal a tissue-specific toxic gain-of-function and mitochondrial stress response. Acta Neuropathol 138, 103–121 (2019). https://doi.org/10.1007/s00401-019-01989-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-01989-y