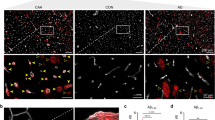
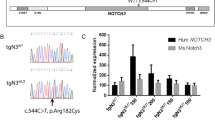
Abstract
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and a phenotypically similar recessive condition (CARASIL) have emerged as important genetic model diseases for studying the molecular pathomechanisms of cerebral small vessel disease (SVD). CADASIL, the most frequent and intensely explored monogenic SVD, is characterized by a severe pathology in the cerebral vasculature including the mutation-induced aggregation of the Notch3 extracellular domain (Notch3ECD) and the formation of protein deposits of insufficiently determined composition in vessel walls. To identify key molecules and pathways involved in this process, we quantitatively determined the brain vessel proteome from CADASIL patient and control autopsy samples (n = 6 for each group), obtaining 95 proteins with significantly increased abundance. Intriguingly, high-temperature requirement protein A1 (HTRA1), the extracellular protease mutated in CARASIL, was found to be strongly enriched (4.9-fold, p = 1.6 × 10−3) and to colocalize with Notch3ECD deposits in patient vessels suggesting a sequestration process. Furthermore, the presence of increased levels of several HTRA1 substrates in the CADASIL proteome was compatible with their reduced degradation as consequence of a loss of HTRA1 activity. Indeed, a comparison with the brain vessel proteome of HTRA1 knockout mice (n = 5) revealed a highly significant overlap of 18 enriched proteins (p = 2.2 × 10−16), primarily representing secreted and extracellular matrix factors. Several of them were shown to be processed by HTRA1 in an in vitro proteolysis assay identifying them as novel substrates. Our study provides evidence for a loss of HTRA1 function as a critical step in the development of CADASIL pathology linking the molecular mechanisms of two distinct SVD forms.






Similar content being viewed by others
References
An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y (2010) Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest Ophthalmol Vis Sci 51:3379–3386. https://doi.org/10.1167/iovs.09-4853
Arboleda-Velasquez JF, Manent J, Lee JH, Tikka S, Ospina C, Vanderburg CR, Frosch MP, Rodriguez-Falcon M, Villen J, Gygi S, Lopera F, Kalimo H, Moskowitz MA, Ayata C, Louvi A et al (2011) Hypomorphic Notch 3 alleles link Notch signaling to ischemic cerebral small-vessel disease. Proc Natl Acad Sci USA 108:E128–135. https://doi.org/10.1073/pnas.1101964108
Baron-Menguy C, Domenga-Denier V, Ghezali L, Faraci FM, Joutel A (2017) Increased Notch3 activity mediates pathological changes in structure of cerebral arteries. Hypertension 69:60–70. https://doi.org/10.1161/HYPERTENSIONAHA.116.08015
Beaufort N, Scharrer E, Kremmer E, Lux V, Ehrmann M, Huber R, Houlden H, Werring D, Haffner C, Dichgans M (2014) Cerebral small vessel disease-related protease HtrA1 processes latent TGF-beta binding protein 1 and facilitates TGF-beta signaling. Proc Natl Acad Sci USA 111:16496–16501. https://doi.org/10.1073/pnas.1418087111
Bonnans C, Chou J, Werb Z (2014) Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15:786–801. https://doi.org/10.1038/nrm3904
Capone C, Cognat E, Ghezali L, Baron-Menguy C, Aubin D, Mesnard L, Stohr H, Domenga-Denier V, Nelson MT, Joutel A (2016) Reducing Timp3 or vitronectin ameliorates disease manifestations in CADASIL mice. Ann Neurol 79:387–403. https://doi.org/10.1002/ana.24573
Capone C, Dabertrand F, Baron-Menguy C, Chalaris A, Ghezali L, Domenga-Denier V, Schmidt S, Huneau C, Rose-John S, Nelson MT, Joutel A (2016) Mechanistic insights into a TIMP3-sensitive pathway constitutively engaged in the regulation of cerebral hemodynamics. Elife 5. https://doi.org/10.7554/eLife.17536
Carare RO, Hawkes CA, Jeffrey M, Kalaria RN, Weller RO (2013) Review: cerebral amyloid angiopathy, prion angiopathy, CADASIL and the spectrum of protein elimination failure angiopathies (PEFA) in neurodegenerative disease with a focus on therapy. Neuropathol Appl Neurobiol 39:593–611. https://doi.org/10.1111/nan.12042
Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG (2009) CADASIL. Lancet Neurol 8:643–653. https://doi.org/10.1016/s1474-4422(09)70127-9
Clausen T, Kaiser M, Huber R, Ehrmann M (2011) HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12:152–162. https://doi.org/10.1038/nrm3065
Cognat E, Baron-Menguy C, Domenga-Denier V, Cleophax S, Fouillade C, Monet-Lepretre M, Dewerchin M, Joutel A (2014) Archetypal Arg169Cys mutation in NOTCH3 does not drive the pathogenesis in cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy via a loss-of-function mechanism. Stroke 45:842–849. https://doi.org/10.1161/STROKEAHA.113.003339
Craggs L, Taylor J, Slade JY, Chen A, Hagel C, Kuhlenbaeumer G, Borjesson-Hanson A, Viitanen M, Kalimo H, Deramecourt V, Oakley AE, Kalaria RN (2016) Clusterin/Apolipoprotein J immunoreactivity is associated with white matter damage in cerebral small vessel diseases. Neuropathol Appl Neurobiol 42:194–209. https://doi.org/10.1111/nan.12248
Craggs LJ, Yamamoto Y, Deramecourt V, Kalaria RN (2014) Microvascular pathology and morphometrics of sporadic and hereditary small vessel diseases of the brain. Brain Pathol 24:495–509. https://doi.org/10.1111/bpa.12177
de la Pena P, Bornstein B, del Hoyo P, Fernandez-Moreno MA, Martin MA, Campos Y, Gomez-Escalonilla C, Molina JA, Cabello A, Arenas J, Garesse R (2001) Mitochondrial dysfunction associated with a mutation in the Notch3 gene in a CADASIL family. Neurology 57:1235–1238
Di Donato I, Bianchi S, De Stefano N, Dichgans M, Dotti MT, Duering M, Jouvent E, Korczyn AD, Lesnik-Oberstein SA, Malandrini A, Markus HS, Pantoni L, Penco S, Rufa A, Sinanovic O et al (2017) Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL) as a model of small vessel disease: update on clinical, diagnostic, and management aspects. BMC Med 15:41. https://doi.org/10.1186/s12916-017-0778-8
Dichgans M, Leys D (2017) Vascular cognitive impairment. Circ Res 120:573–591. https://doi.org/10.1161/CIRCRESAHA.116.308426
Dichgans M, Ludwig H, Muller-Hocker J, Messerschmidt A, Gasser T (2000) Small in-frame deletions and missense mutations in CADASIL: 3D models predict misfolding of Notch3 EGF-like repeat domains. Eur J Hum Genet 8:280–285. https://doi.org/10.1038/sj.ejhg.5200460
Dotti MT, De Stefano N, Bianchi S, Malandrini A, Battisti C, Cardaioli E, Federico A (2004) A novel NOTCH3 frameshift deletion and mitochondrial abnormalities in a patient with CADASIL. Arch Neurol 61:942–945. https://doi.org/10.1001/archneur.61.6.942
Duering M, Karpinska A, Rosner S, Hopfner F, Zechmeister M, Peters N, Kremmer E, Haffner C, Giese A, Dichgans M, Opherk C (2011) Co-aggregate formation of CADASIL-mutant NOTCH3: a single-particle analysis. Hum Mol Genet 20:3256–3265. https://doi.org/10.1093/hmg/ddr237
Ehret F, Vogler S, Pojar S, Elliott DA, Bradke F, Steiner B, Kempermann G (2015) Mouse model of CADASIL reveals novel insights into Notch3 function in adult hippocampal neurogenesis. Neurobiol Dis 75:131–141. https://doi.org/10.1016/j.nbd.2014.12.018
Ghosh M, Balbi M, Hellal F, Dichgans M, Lindauer U, Plesnila N (2015) Pericytes are involved in the pathogenesis of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Ann Neurol 78:887–900. https://doi.org/10.1002/ana.24512
Grau S, Baldi A, Bussani R, Tian X, Stefanescu R, Przybylski M, Richards P, Jones SA, Shridhar V, Clausen T, Ehrmann M (2005) Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc Natl Acad Sci USA 102:6021–6026. https://doi.org/10.1073/pnas.0501823102
Haffner C, Malik R, Dichgans M (2016) Genetic factors in cerebral small vessel disease and their impact on stroke and dementia. J Cereb Blood Flow Metab 36:158–171. https://doi.org/10.1038/jcbfm.2015.71
Hara K, Shiga A, Fukutake T, Nozaki H, Miyashita A, Yokoseki A, Kawata H, Koyama A, Arima K, Takahashi T, Ikeda M, Shiota H, Tamura M, Shimoe Y, Hirayama M et al (2009) Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N Engl J Med 360:1729–1739. https://doi.org/10.1056/NEJMoa0801560
Henshall TL, Keller A, He L, Johansson BR, Wallgard E, Raschperger E, Mae MA, Jin S, Betsholtz C, Lendahl U (2015) Notch3 is necessary for blood vessel integrity in the central nervous system. Arterioscler Thromb Vasc Biol 35:409–420. https://doi.org/10.1161/ATVBAHA.114.304849
Hynes RO (2009) The extracellular matrix: not just pretty fibrils. Science 326:1216–1219. https://doi.org/10.1126/science.1176009
Jones A, Kumar S, Zhang N, Tong Z, Yang JH, Watt C, Anderson J, Amrita Fillerup H, McCloskey M, Luo L, Yang Z, Ambati B, Marc R, Oka C et al (2011) Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc Natl Acad Sci USA 108:14578–14583. https://doi.org/10.1073/pnas.1102853108
Joutel A (2013) Loss-of-function mutation in the NOTCH3 gene: simply a polymorphism? Hum Mutat 34. https://doi.org/10.1002/humu.22198
Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, Piga N, Chapon F, Godfrain C, Tournier-Lasserve E (2000) The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest 105:597–605. https://doi.org/10.1172/jci8047
Joutel A, Haddad I, Ratelade J, Nelson MT (2016) Perturbations of the cerebrovascular matrisome: a convergent mechanism in small vessel disease of the brain? J Cereb Blood Flow Metab 36:143–157. https://doi.org/10.1038/jcbfm.2015.62
Joutel A, Monet-Lepretre M, Gosele C, Baron-Menguy C, Hammes A, Schmidt S, Lemaire-Carrette B, Domenga V, Schedl A, Lacombe P, Hubner N (2010) Cerebrovascular dysfunction and microcirculation rarefaction precede white matter lesions in a mouse genetic model of cerebral ischemic small vessel disease. J Clin Invest 120:433–445. https://doi.org/10.1172/jci39733
Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, Cruaud C, Maciazek J, Weissenbach J, Bousser MG, Bach JF, Tournier-Lasserve E (1997) Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 350:1511–1515. https://doi.org/10.1016/s0140-6736(97)08083-5
Kalaria RN (2016) Neuropathological diagnosis of vascular cognitive impairment and vascular dementia with implications for Alzheimer’s disease. Acta Neuropathol 131:659–685. https://doi.org/10.1007/s00401-016-1571-z
Karbowski M, Neutzner A (2012) Neurodegeneration as a consequence of failed mitochondrial maintenance. Acta Neuropathol 123:157–171. https://doi.org/10.1007/s00401-011-0921-0
Kast J, Hanecker P, Beaufort N, Giese A, Joutel A, Dichgans M, Opherk C, Haffner C (2014) Sequestration of latent TGF-beta binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol Commun 2:96. https://doi.org/10.1186/s40478-014-0096-8
Kofler NM, Cuervo H, Uh MK, Murtomaki A, Kitajewski J (2015) Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL, and results in arteriovenous malformations. Sci Rep 5:16449. https://doi.org/10.1038/srep16449
Lee SJ, Zhang X, Wang MM (2014) Vascular accumulation of the small leucine-rich proteoglycan decorin in CADASIL. NeuroReport 25:1059–1063. https://doi.org/10.1097/WNR.0000000000000230
Lennon FE, Singleton PA (2011) Hyaluronan regulation of vascular integrity. Am J Cardiovasc Dis 1:200–213
Liu C, Xu P, Lamouille S, Xu J, Derynck R (2009) TACE-mediated ectodomain shedding of the type I TGF-beta receptor downregulates TGF-beta signaling. Mol Cell 35:26–36. https://doi.org/10.1016/j.molcel.2009.06.018
Liu H, Zhang W, Kennard S, Caldwell RB, Lilly B (2010) Notch3 is critical for proper angiogenesis and mural cell investment. Circ Res 107:860–870. https://doi.org/10.1161/CIRCRESAHA.110.218271
Machuca-Parra AI, Bigger-Allen AA, Sanchez AV, Boutabla A, Cardona-Velez J, Amarnani D, Saint-Geniez M, Siebel CW, Kim LA, D’Amore PA, Arboleda-Velasquez JF (2017) Therapeutic antibody targeting of Notch3 signaling prevents mural cell loss in CADASIL. J Exp Med 214:2271–2282. https://doi.org/10.1084/jem.20161715
Malandrini A, Albani F, Palmeri S, Fattapposta F, Gambelli S, Berti G, Bracco A, Tammaro A, Calzavara S, Villanova M, Ferrari M, Rossi A, Carrera P (2002) Asymptomatic cores and paracrystalline mitochondrial inclusions in CADASIL. Neurology 59:617–620
Manousopoulou A, Gatherer M, Smith C, Nicoll JAR, Woelk CH, Johnson M, Kalaria R, Attems J, Garbis SD, Carare RO (2017) Systems proteomic analysis reveals that clusterin and tissue inhibitor of metalloproteinases 3 increase in leptomeningeal arteries affected by cerebral amyloid angiopathy. Neuropathol Appl Neurobiol 43:492–504. https://doi.org/10.1111/nan.12342
Megger DA, Bracht T, Meyer HE, Sitek B (2013) Label-free quantification in clinical proteomics. Biochim Biophys Acta 1834:1581–1590. https://doi.org/10.1016/j.bbapap.2013.04.001
Monet-Lepretre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V, Dussaule C, Cognat E, Vinh J, Joutel A (2013) Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain 136:1830–1845. https://doi.org/10.1093/brain/awt092
Muller K, Courtois G, Ursini MV, Schwaninger M (2017) New insight into the pathogenesis of cerebral small-vessel diseases. Stroke 48:520–527. https://doi.org/10.1161/STROKEAHA.116.012888
Nagatoshi A, Ueda M, Ueda A, Tasaki M, Inoue Y, Ma Y, Masuda T, Mizukami M, Matsumoto S, Kosaka T, Kawano T, Ito T, Ando Y (2017) Serum amyloid P component: a novel potential player in vessel degeneration in CADASIL. J Neurol Sci 379:69–76. https://doi.org/10.1016/j.jns.2017.05.033
Nozaki H, Kato T, Nihonmatsu M, Saito Y, Mizuta I, Noda T, Koike R, Miyazaki K, Kaito M, Ito S, Makino M, Koyama A, Shiga A, Uemura M, Sekine Y et al (2016) Distinct molecular mechanisms of HTRA1 mutants in manifesting heterozygotes with CARASIL. Neurology 86:1964–1974. https://doi.org/10.1212/WNL.0000000000002694
Nozaki H, Nishizawa M, Onodera O (2014) Features of cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke 45:3447–3453. https://doi.org/10.1161/strokeaha.114.004236
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144:67–78. https://doi.org/10.1016/j.cell.2010.11.050
Opherk C, Duering M, Peters N, Karpinska A, Rosner S, Schneider E, Bader B, Giese A, Dichgans M (2009) CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum Mol Genet 18:2761–2767. https://doi.org/10.1093/hmg/ddp211
Peters N, Opherk C, Bergmann T, Castro M, Herzog J, Dichgans M (2005) Spectrum of mutations in biopsy-proven CADASIL: implications for diagnostic strategies. Arch Neurol 62:1091–1094. https://doi.org/10.1001/archneur.62.7.1091
Pippucci T, Maresca A, Magini P, Cenacchi G, Donadio V, Palombo F, Papa V, Incensi A, Gasparre G, Valentino ML, Preziuso C, Pisano A, Ragno M, Liguori R, Giordano C et al (2015) Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol Med 7:848–858. https://doi.org/10.15252/emmm.201404399
Ruchoux MM, Domenga V, Brulin P, Maciazek J, Limol S, Tournier-Lasserve E, Joutel A (2003) Transgenic mice expressing mutant Notch3 develop vascular alterations characteristic of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Am J Pathol 162:329–342. https://doi.org/10.1016/S0002-9440(10)63824-2
Rutten JW, Boon EM, Liem MK, Dauwerse JG, Pont MJ, Vollebregt E, Maat-Kievit AJ, Ginjaar HB, Lakeman P, van Duinen SG, Terwindt GM, Lesnik Oberstein SA (2013) Hypomorphic NOTCH3 alleles do not cause CADASIL in humans. Hum Mutat 34:1486–1489. https://doi.org/10.1002/humu.22432
Rutten JW, Dauwerse HG, Peters DJ, Goldfarb A, Venselaar H, Haffner C, van Ommen GB, Aartsma-Rus AM, Lesnik Oberstein SA (2016) Therapeutic NOTCH3 cysteine correction in CADASIL using exon skipping: in vitro proof of concept. Brain 139:123–135. https://doi.org/10.1093/brain/aww011
Rutten JW, Haan J, Terwindt GM, van Duinen SG, Boon EM, Lesnik Oberstein SA (2014) Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev Mol Diagn 14:593–603. https://doi.org/10.1586/14737159.2014.922880
Shiga A, Nozaki H, Yokoseki A, Nihonmatsu M, Kawata H, Kato T, Koyama A, Arima K, Ikeda M, Katada S, Toyoshima Y, Takahashi H, Tanaka A, Nakano I, Ikeuchi T et al (2011) Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-beta1 via cleavage of proTGF-beta1. Hum Mol Genet 20:1800–1810. https://doi.org/10.1093/hmg/ddr063
Sun Y, Vandenbriele C, Kauskot A, Verhamme P, Hoylaerts MF, Wright GJ (2015) A human platelet receptor protein microarray identifies the high affinity immunoglobulin E receptor subunit α (FcεR1α) as an activating platelet endothelium aggregation receptor 1 (PEAR1) ligand. Mol Cell Proteomics 14:1265–1274. https://doi.org/10.1074/mcp.M114.046946
Tan R, Traylor M, Rutten-Jacobs L, Markus H (2017) New insights into mechanisms of small vessel disease stroke from genetics. Clin Sci (Lond) 131:515–531. https://doi.org/10.1042/CS20160825
Tiaden AN, Richards PJ (2013) The emerging roles of HTRA1 in musculoskeletal disease. Am J Pathol 182:1482–1488. https://doi.org/10.1016/j.ajpath.2013.02.003
Tikka S, Baumann M, Siitonen M, Pasanen P, Poyhonen M, Myllykangas L, Viitanen M, Fukutake T, Cognat E, Joutel A, Kalimo H (2014) CADASIL and CARASIL. Brain Pathol 24:525–544. https://doi.org/10.1111/bpa.12181
Verdura E, Herve D, Scharrer E, Amador Mdel M, Guyant-Marechal L, Philippi A, Corlobe A, Bergametti F, Gazal S, Prieto-Morin C, Beaufort N, Le Bail B, Viakhireva I, Dichgans M, Chabriat H et al (2015) Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain 138:2347–2358. https://doi.org/10.1093/brain/awv155
Vierkotten S, Muether PS, Fauser S (2011) Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch’s membrane via cleavage of extracellular matrix components. PLoS One 6:e22959. https://doi.org/10.1371/journal.pone.0022959
Weiming F, Yuliang W, Youjie L, Xinsheng L, Shuyang X, Zhaoxia L (2013) A novel Notch3 deletion mutation in a Chinese patient with cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL). J Clin Neurosci 20:322–323. https://doi.org/10.1016/j.jocn.2012.02.026
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. https://doi.org/10.1038/nmeth.1322
Wollenweber FA, Hanecker P, Bayer-Karpinska A, Malik M, Bäzner H, Moreton F, Muir KW, Müller S, Giese A, Opherk C, Dichgans M, Haffner C, Düring M (2015) Cysteine-sparing CADASIL mutations in NOTCH3 show pro-aggregatory properties in vitro. Stroke 46:786–792
Yang H, Hu HY (2016) Sequestration of cellular interacting partners by protein aggregates: implication in a loss-of-function pathology. FEBS J 283:3705–3717. https://doi.org/10.1111/febs.13722
Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, Sakai S, Sugiyama Y, Enomoto H, Okada Y, Inoue S (2013) KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci USA 110:5612–5617. https://doi.org/10.1073/pnas.1215432110
Yousif S, Marie-Claire C, Roux F, Scherrmann JM, Decleves X (2007) Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res 1134:1–11. https://doi.org/10.1016/j.brainres.2006.11.089
Zhang X, Lee SJ, Young KZ, Josephson DA, Geschwind MD, Wang MM (2014) Latent NOTCH3 epitopes unmasked in CADASIL and regulated by protein redox state. Brain Res 1583:230–236. https://doi.org/10.1016/j.brainres.2014.08.018
Zhang X, Lee SJ, Young MF, Wang MM (2015) The small leucine-rich proteoglycan BGN accumulates in CADASIL and binds to NOTCH3. Transl Stroke Res 6:148–155. https://doi.org/10.1007/s12975-014-0379-1
Acknowledgements
We thank N. Ziesch, B. Lindner and A. Berghofer for excellent technical assistance. This study was supported by the Vascular Dementia Research Foundation (to MD), the Fondation Leducq (Transatlantic Network of Excellence on the Pathogenesis of Small Vessel Disease of the Brain, to MD and AJ), the European Union’s Horizon 2020 research and innovation program (SVDs@target, to MD), the Deutsche Forschungsgemeinschaft (FOR2290, to SL; DI 722/13-1, to MD; HA2448/6-1, to CH), the Helmholtz Israel program (to SL), the Centers of Excellence in Neurodegeneration (to SL), the Breuer foundation Award (to SL) and the National Research Agency of France (ANR-16-RHU RGE16212HKA, to AJ).
Author information
Authors and Affiliations
Contributions
AZ, SM, ES, SL, MD and CH designed the project; SM and SL performed proteomic experiments; AZ and ES performed biochemical experiments; TA, CO, VDD and AJ contributed critical materials and expertise; AZ, SM, ES, SL, MD and CH analyzed data and wrote the manuscript; all authors reviewed and revised the manuscript.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zellner, A., Scharrer, E., Arzberger, T. et al. CADASIL brain vessels show a HTRA1 loss-of-function profile. Acta Neuropathol 136, 111–125 (2018). https://doi.org/10.1007/s00401-018-1853-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1853-8