Abstract
Purpose
Infant gut microbiota which plays an important role in long-term health is mainly shaped by early life nutrition. However, the effect of nutrients on infants gut microbiota is less researched. Here, we present a study aiming to investigate in vitro a modified formula that is supplemented with milk fat globule membrane (MFGM) that were missing in common formulas when compared with human milk and to assess the impact of feeding scheme on microbiota and metabolism.
Methods
A total of 44 infants including 16 from breast milk feeding, 13 from common formula feeding and 15 from modified formula feeding were analyzed, and A cross-sectional sampling of fecal and urine was done at 1 month-of-age. Stool microbiota composition was characterized using high-throughput DNA sequencing, and urinary metabolome was profiled by nuclear magnetic resonance (NMR). In vitro growth experiment of Bifidobacterium with key components from MFGM was performed and analyzed by both DNA and RNA.
Results
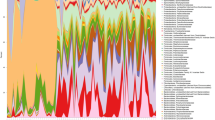
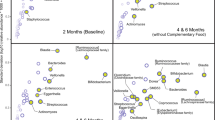
Stool samples from the infants who were breastfed had a higher relative abundance of Bifidobacterium and a lower relative abundance of Escherichia than the formula-fed infants. The stool microbiome shifts were associated with urine metabolites changes. Three substances including lactadherin, sialic acid and phospholipid, key components of MFGM were significantly positively correlated to Bifidobacterium of stool samples from infants, and stimulated the growth rate of Bifidobacterium significantly by provided energy in vitro growth experiment with RNA analysis.
Conclusions
These findings suggest that the key components from MFGM could improve infants’ health by modulating the gut microbiome, and possibly supporting the growth of Bifidobacterium.
Registration
Clinicaltrials.gov NCT02658500 (registered on January 20, 2016).





Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to ethical and privacy considerations but are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Human Microbiome Project Consortium T (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. https://doi.org/10.1038/nature11234
Qin JJ, Li YR, Cai ZM et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. https://doi.org/10.1038/nature11450
Schulz MD, Atay Ç, Heringer J et al (2014) High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514:508–512. https://doi.org/10.1038/nature13398
Ridaura VK, Faith JJ, Rey FE et al (2013) Gut microbiota from twins discordant for obesity modulate metabolism in XZmice. Science 341:1241214. https://doi.org/10.1126/science.1241214
Zimmermann P, Curtis N (2020) Breast milk microbiota: a review of the factors that influence composition. J Infect 81:17–47. https://doi.org/10.1016/j.jinf.2020.01.023
Sindi AS, Geddes DT, Wlodek ME et al (2021) Can we modulate the breastfed infant gut microbiota through maternal diet? FEMS Microbiol Rev. https://doi.org/10.1093/femsre/fuab011
Walker RW, Clemente JC, Peter I, Loos RJF (2017) The prenatal gut microbiome: are we colonized with bacteria in utero ? Pediatr Obes 12:3–17. https://doi.org/10.1111/ijpo.12217
Kostic ADD, Gevers D, Siljander H et al (2015) The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17:260–273. https://doi.org/10.1016/j.chom.2015.01.001
Le Huërou-Luron I, Blat S, Boudry G (2010) Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 23:23–36. https://doi.org/10.1017/S0954422410000065
Guaraldi F, Salvatori G (2012) Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2:1–4. https://doi.org/10.3389/fcimb.2012.00094
Bezirtzoglou E, Tsiotsias A, Welling GW (2011) Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17:478–482. https://doi.org/10.1016/j.anaerobe.2011.03.009
Zhao LL, Du M, Gao J et al (2019) Label-free quantitative proteomic analysis of milk fat globule membrane proteins of yak and cow and identification of proteins associated with glucose and lipid metabolism. Food Chem 275:59–68. https://doi.org/10.1016/j.foodchem.2018.09.04413
Bhinder G, Allaire JM, Garcia C et al (2017) Milk fat globule membrane supplementation in formula modulates the neonatal gut microbiome and normalizes intestinal development. Sci Rep 7:45274. https://doi.org/10.1038/srep45274
Aimee MBD, Alaric WDS, Phillip IT, Barbara BW, Gautam D (2018) Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat med 24:1822–1829. https://doi.org/10.1038/s41591-018-0216-2
Martin CR, Ling PR, Blackburn GL (2016) Review of infant feeding: key features of breast milk and infant formula. Nutrients 8:279. https://doi.org/10.3390/nu8050279
Brink LR, Katelin M, Piccolo BD et al (2019) Neonatal diet impacts bioregional microbiota composition in piglets fed human breast milk or infant formula. J Nutr 149:2236–2246. https://doi.org/10.1093/jn/nxz170
Johnson CC, Ownby DR (2017) The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res 179:60–70. https://doi.org/10.1016/j.trsl.2016.06.010
Stewart CJ, Ajami NJ, O’Brien JL et al (2018) Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 7728:583–588. https://doi.org/10.1038/s41586-018-0617-x
Cukrowska B, Biera JB, Zakrzewska M et al (2020) The Relationship between the infant gut microbiota and allergy. The role of Bifidobacterium breve and prebiotic oligosaccharides in the activation of anti-allergic mechanisms in early life. Nutrients 12:946. https://doi.org/10.3390/nu12040946
Li T, Yang JJ, Zhang HX et al (2020) Bifidobacterium from breastfed infant faeces prevent high-fat-diet-induced glucose tolerance impairment, mediated by the modulation of glucose intake and the incretin hormone secretion axis. J Sci Food Agric 100:3308–3318. https://doi.org/10.1002/jsfa.10360
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2409v1. https://doi.org/10.7717/peerj.2584
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-0
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing. Available from http://www.r-project.org/
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Dray S, Dufour A-BB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Makowski D (2016) Package “neuropsychology”: an R toolbox for psychologists, neuropsychologists and neuroscientists. Available from: https://github.com/neuropsychology/neuropsychology.R Accessed 30 June 2019
Patel RK, Jain M (2012) NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619. https://doi.org/10.1371/journal.pone.0030619
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667. https://doi.org/10.1038/nprot.2016.095
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108–e108. https://doi.org/10.1093/nar/gkt214
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–34. https://doi.org/10.1186/s13059-014-0550-8
Yu G, Wang LG, Han Y, He QY (2012) ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Yu G, Wang LG, Yan GR, He QY (2015) DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31:608–609. https://doi.org/10.1093/bioinformatics/btu684
Cani PD, Hul MV (2020) Mediterranean diet, gut microbiota and health: when age and calories do not add up. Gut 69:1167–1168. https://doi.org/10.1136/gutjnl-2020-320781
Forbes JD, Azad MB, Lorena V et al (2018) Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr 172:e181161. https://doi.org/10.1001/jamapediatrics.2018.1161
Newburg DS, Morelli L (2015) Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res 77:115–120. https://doi.org/10.1038/pr.2014.178
Stearns JC, Zulyniak MA, de Souza RJ et al (2017) Ethnic and diet-related differences in the healthy infant microbiome. Genome Med 9:32. https://doi.org/10.1186/s13073-017-0421-5
Li N, Yan F, Wang N et al (2020) Distinct gut microbiota and metabolite profiles induced by different feeding methods in healthy Chinese infants. Front Microbiol 11:714. https://doi.org/10.3389/fmicb.2020.00714
Thompson AL (2012) Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol 24:350–360. https://doi.org/10.1002/ajhb.22254
Doueellou T, Montel MC, Sergentet DT (2017) Anti-adhesive properties of bovine oligosaccharides and bovine milk fat globule membrane-associated glycoconjugates against bacterial food enteropathogens. J Dairy Sci 100:3348–3359. https://doi.org/10.3168/jds.2016-11611
Underwood MA, German JB, Lebrilla CB, Mills DA (2015) Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res 77:229. https://doi.org/10.1038/pr.2014.156
Bouatra S, Aziat F, Mandal R et al (2013) The human urine metabolome. PLoS ONE 8:e73076. https://doi.org/10.1371/journal.pone.0073076
Bowling FG, Thomas M (2014) Analyzing the metabolome. Methods Mol Biol 1168:31–45. https://doi.org/10.1007/978-1-4939-0847-9_3
Gonthier MP, Cheynier V, Donovan JL, Manach C, Morand C, Mila I, Lapierre C, Rémésy C, Scalbert A (2003) Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J Nutr 133(461–467):42. https://doi.org/10.1046/j.1365-277X.2003.00411.x
O’Sullivan A, He X, McNiven EMS, Hinde K, Haggarty NW, Lönnerdal B, Slupsky CM (2013) Metabolomic phenotyping validates the infant rhesus monkey as a model of human infant metabolism. J Pediatr Gastroenterol Nutr 56:355–363. https://doi.org/10.1097/MPG.0b013e31827e1f07
Riesberg LA, Weed SA, McDonald TL, Eckerson JM, Drescher KM (2016) Beyond muscles: the untapped potential of creatine. Int Immunopharmacol 37:31–42. https://doi.org/10.1016/j.intimp.2015.12.034
Cunha MP, Martin-De-Saavedra MD, Romero A, Javier E, Fabiana KL, Carla IT, Marcelo F, Ana LSR, Manuela GL (2014) Both creatine and its product phosphocreatine reduce oxidative stress and afford neuroprotection in an in vitro Parkinson’s model. ASN Neuro 6:1–6. https://doi.org/10.1177/1759091414554945
Muccioli GG, Naslain D, Bäckhed F, Reigstad CS, Lambert DM, Delzenne NM, Cani PD (2010) The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol 6:392. https://doi.org/10.1038/msb.2010.46
Carlier JP (2015) Veillonella. In: David H (ed) Bergey’s manual of systematics of archaea and bacteria, 2nd edn. John Wiley & Sons, Hoboken, pp 1–11
Acknowledgements
We thank the infants and their parents participating in this study for their trust. We also thank the study team for their involvement in this work, especially nurses and coordinators. We thank Dr Jufang Li and Dr Xueyan Dong for helping collecting samples, and we also thank Dr Jun Wang and Dr Yue Ma for bioinformatic analysis and revision.
Funding
This work was supported by National Natural Science Foundation of China No. 32072191, Beijing Science and Technology Plan No. Z201100008020005, Daxing District Major Scientific and Technological Achievements Transformation Project No. 2020006, National Key Research and Development Program No. 2019YFF0216702, Beijing Science and Technology Plan No. Z201100002620005.
Author information
Authors and Affiliations
Contributions
JZ: investigation, data curation, writing original draft preparation; WY: methodology, investigation, data curation; BL: data curation, writing original draft preparation; YD: methodology, investigation; TJ: methodology, investigation; SC: investigation; JW: investigation; BF: investigation; WQ: investigation; YL: investigation; HZ: investigation; JH: investigation; JH: methodology; LC: conceptualization, methodology, infant formula design, funding acquisition, resources.
Corresponding author
Ethics declarations
Conflict of interest
There was no conflicts of interest/competing interests for this article.
Ethical approval
The Ethics Committee of Beijing Ditan Hospital affiliated to Capital Medical University approved all aspects of the study (#2015-027-01).
Consent to participate
Informed consent was obtained from all of the parents.
Consent for publication
All authors were consent for publicaiton.
Supplementary Information
Below is the link to the electronic supplementary material.
394_2021_2638_MOESM2_ESM.tiff
Supplementary file2 Principle coordinated analysis (unweighted Unifrac distance) of stool microbiome of three feeding schemes including Breast feeding (BF), common formula feeding (FF1) and modified formula feeding (FF2) (TIFF 537 KB)
394_2021_2638_MOESM3_ESM.tif
Supplementary file3 Alpha-diversity analysis of stool microbiome of three feeding schemes. Breast feeding (BF), common formula feeding (FF1) and modified formula feeding (FF2) (TIF 403 KB)
394_2021_2638_MOESM4_ESM.docx
Supplementary file4 Relative abundance of Bifidobacterium, Veillonella and Escherichia/Shigella of three feeding schemes. Breast feeding (BF), common formula feeding (FF1) and modified formula feeding (FF2) (DOCX 1239 KB)
394_2021_2638_MOESM5_ESM.docx
Supplementary file5 Dot plot of gene enrichment analysis of Bifidobacterium cultured with MFGM components in vitro. Supplemental Table 2. Anthropometric measures as well as clinical parameters of 1 month of infant. (DOCX 31 KB)
Rights and permissions
About this article
Cite this article
Zhao, J., Yi, W., Liu, B. et al. MFGM components promote gut Bifidobacterium growth in infant and in vitro. Eur J Nutr 61, 277–288 (2022). https://doi.org/10.1007/s00394-021-02638-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-021-02638-5