Abstract
Purpose
Human breast milk is the optimal source of nutrients for growing infants. However, many circumstances can arise which preclude breast milk feeding, leading to the use of infant formula, including during the weaning period. Many diet-related effects are modulated by the gut microbiome. Therefore, we investigated the effect of human milk (HM) or infant formula (IF) on the gut microbiota in weanling rats.
Methods
The gut microbiota of weanling male Sprague–Dawley rats fed HM or IF for 28 days was analysed by shotgun metagenome sequencing. Caecal contents were analysed by liquid chromatography–mass spectrometry metabolomics.
Results
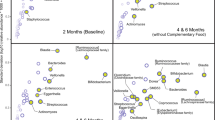
Numerous genera within the Proteobacteria phylum were relatively more abundant in the ileum, caecum, and colon of rats fed HM, including ileal Escherichia (HM = 9.6% ± 4.3 SEM; IF = 0.9% ± 0.3 SEM; P = 0.03). Other taxa that differed between HM- and IF-fed rats included Prevotella and Ruminococcus. Overall, more differences were observed in the ileum than the caecum and colon between rats fed HM and IF. For the rats fed IF, in the ileum, the relative abundance of Bifidobacterium was higher (HM = 1.7% ± 0.7 SEM; IF = 5.0% ± 1.5 SEM; P = 0.04) with gene functions related to carbohydrate and amino acid metabolism also decreased. In the caecum, metabolic features such as bile acids were elevated while amino sugars were also decreased.
Conclusion
Our results show that HM and IF composition differences are reflected in the gut microbiome composition and function in both the small and large intestines.




Similar content being viewed by others
References
Garrido D, Dallas DC, Mills DA (2013) Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology (Reading, England) 159(Pt 4):649–664. https://doi.org/10.1099/mic.0.064113-0
Maga EA, Desai PT, Weimer BC, Dao N, Kultz D, Murray JD (2012) Consumption of lysozyme-rich milk can alter microbial fecal populations. Appl Environ Microbiol 78(17):6153–6160. https://doi.org/10.1128/AEM.00956-12
Maga EA, Weimer BC, Murray JD (2013) Dissecting the role of milk components on gut microbiota composition. Gut Microbes 4(2):136–139. https://doi.org/10.4161/gmic.23188
Macpherson AJ, Uhr T (2004) Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science (New York, NY) 303(5664):1662–1665
Gale C, Logan KM, Santhakumaran S, Parkinson JRC, Hyde MJ, Modi N (2012) Effect of breastfeeding compared with formula feeding on infant body composition: a systematic review and meta-analysis. Am J Clin Nutr 95(3):656–669. https://doi.org/10.3945/ajcn.111.027284
Stuebe A (2009) The risks of not breastfeeding for mothers and infants. Rev Obstet Gynecol 2(4):222–231
Timmerman HM, Rutten NBMM, Boekhorst J, Saulnier DM, Kortman GAM, Contractor N, Kullen M, Floris E, Harmsen HJM, Vlieger AM, Kleerebezem M, Rijkers GT (2017) Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci Rep 7(1):8327. https://doi.org/10.1038/s41598-017-08268-4
Liu Z, Roy NC, Guo Y, Jia H, Ryan L, Samuelsson L, Thomas A, Plowman J, Clerens S, Day L, Young W (2016) Human breast milk and infant formulas differentially modify the intestinal microbiota in human infants and host physiology in rats. J Nutr 146(2):191–199. https://doi.org/10.3945/jn.115.223552
Bengtsson-Palme J, Hartmann M, Eriksson Karl M, Pal C, Thorell K, Larsson Dan Göran J, Nilsson Rolf H (2015) metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15(6):1403–1414. https://doi.org/10.1111/1755-0998.12399
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9:386. https://doi.org/10.1186/1471-2105-9-386
Glass EM, Wilkening J, Wilke A, Antonopoulos D (2010) Meyer F (2010) Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harbor protocols 1:pdb prot5368. https://doi.org/10.1101/pdb.prot5368
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33(17):5691–5702. https://doi.org/10.1093/nar/gki866
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14(6):927–930. https://doi.org/10.1111/j.1654-1103.2003.tb02228.x
Wheeler B, Torchiano M (2016) lmPerm: permutation tests for linear models. R package version 2.1.0.https://CRAN.Rproject.org/package=lmPerm
Subbaraj AK, Kim YHB, Fraser K, Farouk MM (2016) A hydrophilic interaction liquid chromatography–mass spectrometry (HILIC–MS) based metabolomics study on colour stability of ovine meat. Meat Sci 117:163–172
Fraser K, Lane GA, Otter DE, Hemar Y, Quek S-Y, Harrison SJ, Rasmussen S (2013) Analysis of metabolic markers of tea origin by UHPLC and high resolution mass spectrometry. Food Res Int 53(2):827–835. https://doi.org/10.1016/j.foodres.2012.10.015
Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78(3):779–787
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5(3):299–314
Dunn WB, Wilson ID, Nicholls AW, Broadhurst D (2012) The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 4(18):2249–64
Parsons HM, Ekman DR, Collette TW, Viant MR (2009) Spectral relative standard deviation: a practical benchmark in metabolomics. Analyst 134(3):478–485
Xia J, Sinelnikov IV, Han B, Wishart DS (2015) MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic acids research 43(W1):W251–W257
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3(3):211–221
Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27(6):747–751
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45(7):703–714
Romano KA, Vivas EI, Amador-Noguez D, Rey FE (2015) Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6(2):e02481. https://doi.org/10.1128/mBio.02481-14
Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG (2014) Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA 111(20):7421–7426. https://doi.org/10.1073/pnas.1323599111
Hagey LR, Krasowski MD (2013) Microbial biotransformations of bile acids as detected by electrospray mass spectrometry. Adv Nutr Int Rev J 4(1):29–35
Barroso LA, Dean J, Holzle U (2003) Web search for a planet: the Google cluster architecture. IEEE Micro 23(2):22–28
Masarwi M, Solnik HI, Phillip M, Yaron S, Shamir R, Pasmanic-Chor M, Gat-Yablonski G (2018) Food restriction followed by refeeding with a casein- or whey-based diet differentially affects the gut microbiota of pre-pubertal male rats. J Nutr Biochem 51:27–39. https://doi.org/10.1016/j.jnutbio.2017.08.014
Pokusaeva K, Fitzgerald GF, van Sinderen D (2011) Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6(3):285–306. https://doi.org/10.1007/s12263-010-0206-6
Mengheri E, Nobili F, Vignolini F, Pesenti M, Brandi G, Biavati B (1999) Bifidobacterium animalis protects intestine from damage induced by zinc deficiency in rats. J Nutr 129(12):2251–2257. https://doi.org/10.1093/jn/129.12.2251
Aoki R, Tsuchida S, Arai Y, Ohno K, Nishijima T, Mawatari T, Mikami Y, Ushida K (2016) Effect of Bifidobacterium animalis subsp. lactis GCL2505 on the physiological function of intestine in a rat model. Food Sci Nutr 4(6):782–790. https://doi.org/10.1002/fsn3.344
Chen MF, Weng KF, Huang SY, Liu YC, Tseng SN, Ojcius DM, Shih SR (2016) Pretreatment with a heat-killed probiotic modulates monocyte chemoattractant protein-1 and reduces the pathogenicity of influenza and enterovirus 71 infections. Mucosal Immunol 10:215. https://doi.org/10.1038/mi.2016.31
Ulluwishewa D, Anderson RC, Young W, McNabb WC, van Baarlen P, Moughan PJ, Wells JM, Roy NC (2015) Live Faecalibacterium prausnitzii in an apical anaerobic model of the intestinal epithelial barrier. Cell Microbiol 17(2):226–240. https://doi.org/10.1111/cmi.12360
Bode L (2012) Human Milk Oligosaccharides: every baby needs a sugar mama. Glycobiology. https://doi.org/10.1093/glycob/cws074
Bode L (2018) Human milk oligosaccharides in the prevention of necrotizing enterocolitis: a journey from in vitro and in vivo models to mother-infant cohort studies. Front Pediatr 6(385):10. https://doi.org/10.3389/fped.2018.00385
Luchansky SJ, Yarema KJ, Takahashi S, Bertozzi CR (2003) GlcNAc 2-epimerase can serve a catabolic role in sialic acid metabolism. J Biol Chem 278(10):8035–8042
Ridlon JM, Kang DJ, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47(2):241–259. https://doi.org/10.1194/jlr.R500013-JLR200
Hammons J, Jordan W, Stewart R, Taulbee J, Berg R (1988) Age and diet effects on fecal bile acids in infants. J Pediatr Gastroenterol Nutr 7(1):30–38
Mercer KE, Diaz-Rubio ME, Bhattacharyya S, Sharma N, Yeruva L (2016) Programming effects of infant diet on cholesterol/bile acid synthesis and absorption in piglets. FASEB J 30(1_supplement):267.266. https://doi.org/10.1096/fasebj.30.1_supplement.267.6
Halpern MD, Dvorak B (2008) Does abnormal bile acid metabolism contribute to NEC? Seminars in perinatology, vol 2. Elsevier, Amsterdam, pp 114–121
Garrido D, Ruiz-Moyano S, Mills DA (2012) Release and utilization of N-acetyl-d-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe 18(4):430–435. https://doi.org/10.1016/j.anaerobe.2012.04.012
Chen T, Long W, Zhang C, Liu S, Zhao L, Hamaker BR (2017) Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep 7(1):2594. https://doi.org/10.1038/s41598-017-02995-4
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107(33):14691–14696. https://doi.org/10.1073/pnas.1005963107
Ze X, Duncan SH, Louis P, Flint HJ (2012) Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6(8):1535–1543. https://doi.org/10.1038/ismej.2012.4
Coppa GV, Gabrielli O, Zampini L, Galeazzi T, Ficcadenti A, Padella L, Santoro L, Soldi S, Carlucci A, Bertino E, Morelli L (2011) Oligosaccharides in 4 different milk groups, Bifidobacteria, and Ruminococcus obeum. J Pediatr Gastroenterol Nutr 53(1):80–87. https://doi.org/10.1097/MPG.0b013e3182073103
Fuhrer A, Sprenger N, Kurakevich E, Borsig L, Chassard C, Hennet T (2010) Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med 207(13):2843–2854. https://doi.org/10.1084/jem.20101098
Acknowledgements
We would like to acknowledge the technical assistance of Hedley Stirrat for the metabolomics analysis and Paul Maclean for bioinformatics support. All authors designed the study; WY wrote the manuscript with input from all authors; ZL, AS, KF, HJ, WC, LD, NCR, and WY conducted the research and analysed the data; WY, NCR, LD had primary responsibility for the final content. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This study was funded by Bright Dairy & Food Co. Ltd. Zhenmin Liu, Hongxin Jia, and Wenliang Chen are employees of Bright Dairy & Food Co. Ltd. Bright Dairy & Food Co. Ltd is a producer of infant formulae; Arvind Subbaraj, Karl Fraser, Li Day, Nicole, and Wayne Young declare no conflict of interests.
Ethics
Collection of HM was performed with approved consent by participants, Approval no. XHEC-C-2012-024, Xinhua Hospital Ethics Committee Affiliated to Shanghai Jiào tong University School of Medicine. The animal study was conducted under the oversight of the AgResearch Grasslands Animal Ethics Committee (approval number AEC13099; Palmerston North, New Zealand) in accordance with the New Zealand Animal Welfare Act 1999.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Z., Subbaraj, A., Fraser, K. et al. Human milk and infant formula differentially alters the microbiota composition and functional gene relative abundance in the small and large intestines in weanling rats. Eur J Nutr 59, 2131–2143 (2020). https://doi.org/10.1007/s00394-019-02062-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-019-02062-w