Abstract
Purpose
Walnut phenolic extract (WPE) reduces proliferation and enhances differentiation of colon cancer stem cells (CSCs). The present study investigated the metabolic influence of WPE on the mitochondrial function of colon CSCs to determine its underlying mechanism.
Methods
CD133+CD44+ HCT116 colon cancer cells were selected by fluorescence-activated cell sorting and were treated with or without 40 µg/mL WPE. RNA-sequencing (RNA-Seq) was performed to identify differentially expressed genes (DEGs), which were further validated with RT-PCR. WPE-induced alterations in mitochondrial function were investigated through a mitochondrial stress test by determining cellular oxygen consumption rate (OCR), an indicator of mitochondrial respiration, and extracellular acidification rate (ECAR), an indicator of glycolysis, which were further confirmed by glucose uptake and lactate production tests.
Results
RNA-Seq analysis identified two major functional clusters: metabolic and mitochondrial clusters. WPE treatment shifted the metabolic profile of cells towards the glycolysis pathway (ΔECAR = 36.98 mpH/min/ptn, p = 0.02) and oxidative pathway (ΔOCR = 29.18 pmol/min/ptn, p = 0.00001). Serial mitochondrial stimulations using respiration modulators, oligomycin, carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone, and rotenone/antimycin A, found an increased potential of mitochondrial respiration (ΔOCR = 111.5 pmol/min/ptn, p = 0.0006). WPE treatment also increased glucose uptake (Δ = 0.39 pmol/µL, p = 0.002) and lactate production (Δ = 0.08 nmol/µL, p = 0.005).
Conclusions
WPE treatment shifts the mitochondrial metabolism of colon CSC towards more aerobic glycolysis, which might be associated with the alterations in the characteristics of colon CSC.





Similar content being viewed by others
Abbreviations
- WPE:
-
Walnut phenolic extracts
- CSC:
-
Cancer stem cell
- RNA-Seq:
-
RNA-sequencing
- FACS:
-
Fluorescence-activated cell sorting
- DEGs:
-
Differentially expressed genes
- OCR:
-
Oxygen consumption rate
- ECAR:
-
Extracellular acidification rate
- 2-DG:
-
2-Deoxyglucose
- GO:
-
Gene ontology
- DAVID:
-
The Database for Annotation, Visualization and Integrated Discovery
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- FCCP:
-
Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone
References
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Warburg O (1956) On the origin of cancer cells. Science 123(3191):309–314
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23(1):27–47
Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP (2016) Cancer stem cell metabolism. BCR 18(1):55
Fernandez-Arroyo S, Cuyas E, Bosch-Barrera J, Alarcon T, Joven J, Menendez JA (2015) Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience 2(12):958–967
Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J (2010) The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28(4):721–733
Facucho-Oliveira JM JCSJ (2009) The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep 5:140
St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, Schatten GP (2005) The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells 7(3):141–153
Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK (2006) Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun 348(4):1472–1478
Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC (2007) Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci 120(Pt 22):4025–4034
Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH (2015) Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 14(1):86–98
Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14(2):264–271
Hardman WE, Ion G (2008) Suppression of implanted MDA-MB 231 human breast cancer growth in nude mice by dietary walnut. Nutr Cancer 60(5):666–674
Reiter RJ, Tan DX, Manchester LC, Korkmaz A, Fuentes-Broto L, Hardman WE, Rosales-Corral SA, Qi W (2013) A walnut-enriched diet reduces the growth of LNCaP human prostate cancer xenografts in nude mice. Cancer Investig 31(6):365–373
Tsoukas MA, Ko BJ, Witte TR, Dincer F, Hardman WE, Mantzoros CS (2015) Dietary walnut suppression of colorectal cancer in mice: mediation by miRNA patterns and fatty acid incorporation. J Nutr Biochem 26(7):776–783
Lee J, Kim YS, Heo SC, Lee KL, Choi SW, Kim Y (2016) Walnut phenolic extract and its bioactive compounds suppress colon cancer cell growth by regulating colon cancer stemness. Nutrients 8(7):439
Anderson KJ, Teuber SS, Gobeille A, Cremin P, Waterhouse AL, Steinberg FM (2001) Walnut polyphenolics inhibit in vitro human plasma and LDL oxidation. J Nutr 131(11):2837–2842
Min SJ, Lim JY, Kim HR, Kim SJ, Kim Y (2015) Sasa quelpaertensis leaf extract inhibits colon cancer by regulating cancer cell stemness in vitro and in vivo. Int J Mol Sci 16(5):9976–9997
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25(9):1105–1111
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578
Bindea G, Galon J, Mlecnik B (2013) CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29(5):661–663
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J (2009) ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8):1091–1093
Arduini A, Serviddio G, Escobar J, Tormos AM, Bellanti F, Vina J, Monsalve M, Sastre J (2011) Mitochondrial biogenesis fails in secondary biliary cirrhosis in rats leading to mitochondrial DNA depletion and deletions. Am J Physiol Gastrointest Liver Physiol 301(1):G119–G127
Menendez JA (2015) Metabolic control of cancer cell stemness: lessons from iPS cells. Cell Cycle 14(24):3801–3811
Ramm Sander P, Hau P, Koch S, Schutze K, Bogdahn U, Kalbitzer HR, Aigner L (2013) Stem cell metabolic and spectroscopic profiling. Trends Biotechnol 31(3):204–213
Dando I, Dalla Pozza E, Biondani G, Cordani M, Palmieri M, Donadelli M (2015) The metabolic landscape of cancer stem cells. IUBMB Life 67(9):687–693
Kraft CS, LeMoine CM, Lyons CN, Michaud D, Mueller CR, Moyes CD (2006) Control of mitochondrial biogenesis during myogenesis. Am J Physiol Cell Physiol 290(4):C1119–C1127
Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH (2008) Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26(4):960–968
Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, Sugihara E, Onishi N, Yamamoto T, Yanagawa H, Suematsu M, Saya H (2012) Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res 72(6):1438–1448
Li W, Cohen A, Sun Y, Squires J, Braas D, Graeber TG, Du L, Li G, Li Z, Xu X, Chen X, Huang J (2016) The role of CD44 in glucose metabolism in prostatic small cell neuroendocrine carcinoma. MCR 14(4):344–353
Chen KY, Liu X, Bu P, Lin CS, Rakhilin N, Locasale JW, Shen X (2014) A metabolic signature of colon cancer initiating cells. In: Conference proceedings: annual international conference of the IEEE engineering in medicine and biology society IEEE engineering in medicine and biology society annual conference 2014, pp 4759–4762
Krauss S, Zhang CY, Lowell BB (2005) The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol 6(3):248–261
Wang Y, Huang L, Abdelrahim M, Cai Q, Truong A, Bick R, Poindexter B, Sheikh-Hamad D (2009) Stanniocalcin-1 suppresses superoxide generation in macrophages through induction of mitochondrial UCP2. J Leukoc Biol 86(4):981–988
Ricquier D, Casteilla L, Bouillaud F (1991) Molecular studies of the uncoupling protein. FASEB J 5(9):2237–2242
Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A, Thangaraj K, Buchsbaum DJ, LoBuglio AF, Singh KK (2011) Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PloS One 6(9):e24792
Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, Jung HJ, McCaffery JM, Kurland IJ, Reue K, Lee WN, Koehler CM, Teitell MA (2011) UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 30(24):4860–4873
Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, Scarcia P, Palmieri F, Bouillaud F, Fiermonte G (2014) UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci USA 111(3):960–965
Ding S, Li C, Cheng N, Cui X, Xu X, Zhou G (2015) Redox regulation in cancer stem cells. Oxid Med Cell Longev 2015:750–798
Mody N, Parhami F, Sarafian TA, Demer LL (2001) Oxidative stress modulates osteoblastic differentiation of vascular and bone cells. Free Radic Biol Med 31(4):509–519
Kaim G, Dimroth P (1999) ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J 18(15):4118–4127
Sanderson TH, Reynolds CA, Kumar R, Przyklenk K, Huttemann M (2013) Molecular mechanisms of ischemia-reperfusion injury in brain: pivotal role of the mitochondrial membrane potential in reactive oxygen species generation. Mol Neurobiol 47(1):9–23
Acknowledgements
This work was supported by the California Walnut Commission. All authors have participated in the conception, design, and conduction of the study, as well as interpretation of data and drafting the manuscript. J. C. and P. K. S. also performed cell culture studies and measured endpoints. C. H. conducted RNA-Seq and data analysis. Y. K. and S. W. C. have supervised the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
394_2018_1708_MOESM1_ESM.pptx
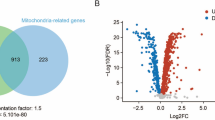
Supplementary material 1. The volcano plot that represents the overall changes in the transcription by WPE treatment. Log2 fold changes of WPE/Ctrl were plotted against –log10 p value. Red dots represent significantly differentially expressed transcripts (p < 0.05, fold change ≥ 1.5). WPE: WPE-treated group. Ctrl: Control group (PPTX 103 KB)
394_2018_1708_MOESM2_ESM.pptx
Supplementary material 2. Functional enrichment analysis and cluster distribution network of up- and down-regulated genes in the WPE-treated CD133+CD44+ HCT116 cells. The circular nodes indicate the ontology terms and pathways of DEGs, which are functionally grouped and interconnected based on the kappa score. The terms and pathways with adjusted p values less than 0.05 were selected for the network construction. The size of the nodes represents the term significance after Bonferroni correction. The significant terms of each group are highlighted. (A) A cluster distribution network of enriched functions and pathways of the 1168 upregulated genes, which are categorized into four major clusters: cluster1 (metabolism), cluster2 (mitochondrion), cluster3 (apoptosis), and cluster4 (cancer pathway), (B) A cluster distribution network of the 815 downregulated genes are representing four major clusters: cluster1 (DNA metabolic process), cluster2 (mitotic cell cycle), cluster3 (RNA metabolic process), and cluster4 (centriole). Both figures are generated by the ClueGO and CluePedia plugin (PPTX 604 KB)
Rights and permissions
About this article
Cite this article
Choi, J., Shin, PK., Kim, Y. et al. Metabolic influence of walnut phenolic extract on mitochondria in a colon cancer stem cell model. Eur J Nutr 58, 1635–1645 (2019). https://doi.org/10.1007/s00394-018-1708-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-018-1708-z