Abstract
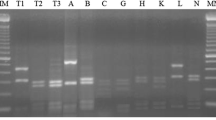
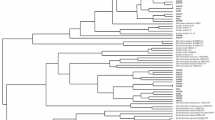
The present study was conducted to isolate and characterize rhizobial strains from root nodules of cultivated legumes, i.e. chickpea, mungbean, pea and siratro. Preliminary characterization of these isolates was done on the basis of plant infectivity test, acetylene reduction assay, C-source utilization, phosphate solubilization, phytohormones and polysaccharide production. The plant infectivity test and acetylene reduction assay showed effective root nodule formation by all the isolates on their respective hosts, except for chickpea isolate Ca-18 that failed to infect its original host. All strains showed homology to a typical Rhizobium strain on the basis of growth pattern, C-source utilization and polysaccharide production. The strain Ca-18 was characterized by its phosphate solubilization and indole acetic acid (IAA) production. The genetic relationship of the six rhizobial strains was carried out by random amplified polymorphic DNA (RAPD) including a reference strain of Bradyrhizobium japonicum TAL-102. Analysis conducted with 60 primers discriminated between the strains of Rhizobium and Bradyrhizobium in two different clusters. One of the primers, OPB-5, yielded a unique RAPD pattern for the six strains and well discriminated the non-nodulating chickpea isolate Ca-18 from all the other nodulating rhizobial strains. Isolate Ca-18 showed the least homology of 15% and 18% with Rhizobium and Bradyrhizobium, respectively, and was probably not a (Brady)rhizobium strain. Partial 16S rRNA gene sequence analysis for MN-S, TAL-102 and Ca-18 strains showed 97% homology between MN-S and TAL-102 strains, supporting the view that they were strains of B. japonicum species. The non-infective isolate Ca-18 was 67% different from the other two strains and probably was an Agrobacterium strain.



Similar content being viewed by others
References
Anand RC, Dogra RC (1991) Physiological and biochemical characteristics of fast and slow growing Rhizobium sp. pigeonpea (Cajanus cajan). J Appl Bacteriol 70:197–202
Barnet Y, Catt PC, Jenzaniontham R, Mann K (1993) Fast growing root nodule bacteria from Australian Acacia spp. In: Palacios R, Mora J, Newton WE (eds) New horizons in nitrogen fixation. Kluwer, Dordrecht, p 594
Bashan Y, Holguin G (1997) Azospirillum—plant relationships: environmental and physiological advances (1990–1996). Can J Microbiol 43:103–121
Bowdich BM, Albright DG, Williams J, Braun MJ (1993) The use of RAPD markers in comparative genome studies. Methods Enzymol 224:294–309
Del Papa MF, Balague LJ, Sowinski SC, Wegener C, Segundo E, Abarca FM, Toro N, Niehaus K, Puhler A, Aguilar OM, Martinez-Drets G, Lagares A (1999) Isolation and characterization of alfalfa-nodulating rhizobia present in acidic soil of central Argentina and Uruguay. Appl Environ Microbiol 65:1420–1427
Echeverrigaray S, Toreson SP, Carrau JL (2000) RAPD marker polymorphism among commercial winery yeast strains. World J Microbiol Biotech 16:143–146
Gordon SA, Weber RP (1951) Colorimetric estimation of indole acetic acid. Plant Physiol 26:192–195
Gougeon AJ, Jobert SD, Shacoori ZT, Menard C, Cormier M (2000) Osmotic stress-induced genetic rearrangements in Escherichia coli H 10407 detected by randomly amplified polymorphic DNA analysis. Appl Environ Microbiol 66:435–438
Hafeez FY, Asad S, Ahmad T, Malik KA (1995) Host specificity and characterization of fast growing rhizobia from Macroptilium atropurpureum cv. siratro in Pakistan. Soil Biol Biochem 27:729–733
Hameed S, Hafeez FY, Mirza MS, Malik KA, Akkermans ADL (1994) Confirmation of an isolate from Datisca cannabina as atypical Frankia strain using PCR amplified 16SrRNA sequence analysis. Pak J Bot 26:247–251
Hardy RWF, Holsten RD, Jackson EK, Burns RE (1968) The acetylene-ethylene assay for nitrogen fixation: laboratory and field evaluation. Plant Physiol 43:1185–1207
Hollis AB, Kloos AB, Elkan GH (1981) DNA: DNA hybridization studies of Rhizobium japonicum and Rhizobiaceae. J Gen Microbiol 123:215–222
Keyser HH, Bohlool BB, Weber DF (1982) Fast growing rhizobia isolated from root nodules of soybean. Science 215:1631–1632
Liu WT, Marsh TL, Cheng H, Forney JL (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of gene encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbour, N.Y.
Martinez-Romero E (1994) Recent development in Rhizobium taxonomy. Plant Soil 161:11–20
Miller GL (1959) Use of dinitrosalicylic acid reagent for the determination of reducing sugars. Anal Chem 31:426–428
Mirza MS, Hameed S, Akkermans ADL (1994) Genetic diversity of Datisca cannabina compatible Frankia strains as determined by sequence analysis of PCR amplified 16S rRNA gene. Appl Environ Microbiol 7:2371–2376
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuweenhoek J Microbiol Serol 73:127–141
Nei N, Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonuclease. Proc Natl Acad Sci USA 76:5269–5273
Nelson LM, Child JJ (1981) Nitrogen fixation and hydrogen metabolism by Rhizobium leguminosarum isolates in pea root nodules. Can J Microbiol 27:1028–1034
Okon Y, Albercht SL, Burris RH (1977) Methods for growing Spirillum lipoferum and for counting it in pure culture and in association with plants. Appl Environ Microbiol 33:85–88
Paffetti D, Scotti C, Gnocchi S, Fancelli S, Bazzicaupo M (1996) Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl Environ Microbiol 62:2279–2285
Pikovskaia RI (1948) Metabolism of phosphorous in soil in connection with vital activity of some microbial species. Microbiologia 17:362–370
Rennie RJ (1981) A single medium for the isolation of nitrogen fixing bacteria. Can J Microbiol 27:8–14
Saleena ML, Loganathan P, Rangarajan S (2001) Genetic diversity and relationship between Bradyrhizobium strains isolated from black gram and cowpea. Biol Fertil Soils 34:276–281
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Sardar R (2000) Growth hormone production by Rhizobium species and their effect on plant growth and nitrogen fixation. M.Phil. thesis, Department of Chemistry, University of Agriculture, Faisalabad, Pakistan
Selenska-Pobell S, Gigova L, Petrova N (1995) Strain specific fingerprints of Rhizobium galegae generated by PCR with arbitrary and repetitive primers. Appl Environ Microbiol 79:425–431
Selenska-Pobell S, Evguenieva-Hacenberg E, Radeva G, Squartini A (1996) Characterization of Rhizobium ‘hedysari’ by RFLP analysis of PCR amplified rDNA and by genomic PCR fingerprinting. J Appl Bacteriol 80:517–528
Sprent JI (1994) Evolution and diversity in the legume-rhizobium symbiosis: chaos theory? Plant Soil 161:1–10
Stanley J, Brown GG, Verma DPS (1985) Studying Rhizobium japonicum comprises two highly diverse symbiotic types. J Bacteriol 163:148–151
Terefework Z, Paulin L, Suomalainen S, Nick G, Lindstrom K (1998) Phylogeny of Rhizobium galegae compared with other rhizobia and agrobacteria. Int J Syst Bacteriol 48:349–350
Tien TM, Gaskins MH, Hubbell DH (1979) Plant growth substances produced by Azospirillum brazilense and their effect on the growth of pearl millet. Appl Environ Microbiol 37:1016–1024
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. International biological program hand book no.15. Blackwell, Oxford, pp 73–97
Williams JGK, Kubelic AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Woese CR, Stackebrand E, Mache T, Fox GE (1985) A definition of the eu-bacterial divisions. Syst Appl Microbiol 6:143–151
Wu KS, Tanksley SD (1993) Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Genet 241:225–235
Yoshida S, Forno DA, Cock JH, Gomex KA (1976) Manual of physiological studies of rice. IRRI, Osbanos, Philippines
Young DC, Oryaizu Y, Oyaizu H, Olsen GJ, Woese ER (1985) Mitochondrial origins. Proc Natl Acad Sci USA 82:4443–4447
Young JPW, Downer HL, Eardly D (1991) Phylogeny of phototrophic Rhizobium strain, BTail by polymerase chain reaction based sequencing of 16SrRNA gene segment. J Bacteriol 173:2271–2277
Acknowledgements
The research work was funded partly by the IDB BIRCEN project and FAO/IAEA TC project no. PAK/5/037. We are thankful to Ms. Fathia Mubeen and Mr. Mazhar Hussain for their help in DNA sequencing work, as well as to Dr. Farooq Latif for his help with HPLC to detect phytohormones.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hameed, S., Yasmin, S., Malik, K.A. et al. Rhizobium, Bradyrhizobium and Agrobacterium strains isolated from cultivated legumes. Biol Fertil Soils 39, 179–185 (2004). https://doi.org/10.1007/s00374-003-0697-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-003-0697-z