Abstract
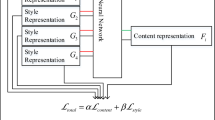
Cervical cell image styles may vary due to factors such as specimen preparation methods and staining schemes. These variations can cause inconsistencies among pathologists and degrade the model performance. Existing staining standardization networks often fail to achieve structure preservation and style approximation. We propose a style-aware network (SA-Net) for multi-domain image normalization to address this issue. SA-Net incorporates a style perception task into the CycleGAN generator to identify different image styles, thus avoiding the need for multiple generators in real-world applications. additionally, We also employ pixel-wise convolutional kernels in the generator to learn only the image style and preserve the image structure. Our experiments demonstrate that SA-Net can effectively enhance the model’s generalization ability and outperform the state-of-the-art methods in multi-style standardization.







Similar content being viewed by others
Data availability
Due to the nature of this research, participants of this study did not agree for their data to be shared publicly, so supporting data is not available.
References
BenTaieb, A., Hamarneh, G.: Adversarial stain transfer for histopathology image analysis. IEEE Trans. Med. Imaging 37(3), 792–802 (2017)
Chen, X., Yu, J., Cheng, S., Geng, X., Liu, S., Han, W., Hu, J., Chen, L., Liu, X., Zeng, S.: An unsupervised style normalization method for cytopathology images. Comput. Struct. Biotechnol. J. 19, 3852–3863 (2021)
Choi, Y., Choi, M., Kim, M., Ha, J.-W., Kim, S., Choo, J.: Stargan: unified generative adversarial networks for multi-domain image-to-image translation. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8789–8797 (2018)
Cong, C., Liu, S., Di Ieva, A., Pagnucco, M., Berkovsky, S., Song, Y.: Texture enhanced generative adversarial network for stain normalisation in histopathology images. In 2021 IEEE 18th international symposium on biomedical imaging (ISBI). IEEE, pp. 1949–1952 (2021)
Cong, C., Liu, S., Di Ieva, A., Pagnucco, M., Berkovsky, S., Song, Y.: Colour adaptive generative networks for stain normalisation of histopathology images. Med. Image Anal. 82, 102580 (2022)
Hetz, M.J., Bucher, T.-C., Brinker, T.J.: Multi-domain stain normalization for digital pathology: a cycle-consistent adversarial network for whole slide images. arXiv preprint arXiv:2301.09431 (2023)
Hoque, M.Z., Keskinarkaus, A., Nyberg, P., Seppänen, T.: Retinex model based stain normalization technique for whole slide image analysis. Comput. Med. Imaging Graph. 90, 101901 (2021)
Isola, P., Zhu, J.-Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017)
Kang, H., Luo, D., Chen, L., Hu, J., Cheng, S., Quan, T., Zeng, S., Liu, X.: Paramnet: A parameter-variable network for fast stain normalization. arXiv preprint arXiv:2305.06511 (2023)
Lei, G., Xia, Y., Zhai, D.-H., Zhang, W., Chen, D., Wang, D.: Staincnns: an efficient stain feature learning method. Neurocomputing 406, 267–273 (2020)
Liang, M., Zhang, Q., Wang, G., Xu, N., Wang, L., Liu, H., Zhang, C.: Multi-scale self-attention generative adversarial network for pathology image restoration. Vis. Comput. 39(9), 4305–4321 (2023)
Ling, Y., Tan, W., Yan, B.: Self-supervised digital histopathology image disentanglement for arbitrary domain stain transfer. IEEE Trans. Med. Imaging (2023)
Lo, Y.-C., Chung, I.-F., Guo, S.-N., Wen, M.-C., Juang, C.-F.: Cycle-consistent gan-based stain translation of renal pathology images with glomerulus detection application. Appl. Soft Comput. 98, 106822 (2021)
Lu, N., Chen, Y.: Multi-category domain-dependent feature-based medical image translation. Visual Comput., 1–20 (2023)
Mahapatra, D., Bozorgtabar, B., Thiran, J.-P., Shao, L.: Structure preserving stain normalization of histopathology images using self supervised semantic guidance. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 309–319 (2020)
Mahapatra, S., Maji, P.: Truncated normal mixture prior based deep latent model for color normalization of histology images. IEEE Trans. Med. Imaging (2023)
Nanni, L., Paci, M., Brahnam, S., Lumini, A.: Comparison of different image data augmentation approaches. J. Imaging 7(12), 254 (2021)
Nazki, H., Arandjelović, O., Um, I., Harrison, D.: Multipathgan: structure preserving stain normalization using unsupervised multi-domain adversarial network with perception loss. arXiv preprint arXiv:2204.09782 (2022)
Nishar, H., Chavanke, N., Singhal, N.: Histopathological stain transfer using style transfer network with adversarial loss. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 330–340 (2020)
Patil, A., Talha, M., Bhatia, A., Kurian, N.C., Mangale, S., Patel, S., Sethi, A.: Fast, self supervised, fully convolutional color normalization of H &E stained images. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). IEEE, pp. 1563–1567 (2021)
Pérez-Bueno, F., Serra, J.G., Vega, M., Mateos, J., Molina, R., Katsaggelos, A.K.: Bayesian k-svd for h and e blind color deconvolution. applications to stain normalization, data augmentation and cancer classification. Comput. Med. Imaging Graphics 97, 102048 (2022)
Plass, M., Kargl, M., Nitsche, P., Jungwirth, E., Holzinger, A., Müller, H.: Understanding and explaining diagnostic paths: toward augmented decision making. IEEE Comput. Graphics Appl. 42(6), 47–57 (2022)
Shaban, M.T., Baur, C., Navab, N., Albarqouni, S.: Staingan: stain style transfer for digital histological images. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (Isbi 2019). IEEE, pp. 953–956 (2019)
Shen, Y., Luo, Y., Shen, D., Ke, J.: Randstainna: learning stain-agnostic features from histology slides by bridging stain augmentation and normalization. In: International Conference on Medical Image Computing and Computer-assisted Intervention. Springer, pp. 212–221 (2022)
Tellez, D., Litjens, G., Bándi, P., Bulten, W., Bokhorst, J.-M., Ciompi, F., Van Der Laak, J.: Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019)
Tosta, T.A.A., de Faria, P.R., Servato, J.P.S., Neves, L.A., Roberto, G.F., Martins, A.S., do Nascimento, M.Z.: Unsupervised method for normalization of hematoxylin-eosin stain in histological images. Comput. Med. Imaging Graph. 77, 101646 (2019)
Tosta, T.A.A., Freitas, A.D., de Faria, P.R., Neves, L.A., Martins, A.S., do Nascimento, M. Z.,: A stain color normalization with robust dictionary learning for breast cancer histological images processing. Biomed. Signal Process. Control 85, 104978 (2023)
Vijh, S., Saraswat, M., Kumar, S.: A new complete color normalization method for h &e stained histopatholgical images. Appl. Intell. 51(11), 7735–7748 (2021)
Wagner, S.J., Khalili, N., Sharma, R., Boxberg, M., Marr, C., Back, W.D., Peng, T.: Structure-preserving multi-domain stain color augmentation using style-transfer with disentangled representations. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 257–266 (2021)
Ye, H.-L., Wang, D.-H.: Stain-adaptive self-supervised learning for histopathology image analysis. arXiv preprint arXiv:2208.04017 (2022)
Zhao, B., Han, C., Pan, X., Lin, J., Yi, Z., Liang, C., Chen, X., Li, B., Qiu, W., Li, D., et al.: Restainnet: a self-supervised digital re-stainer for stain normalization. Comput. Electr. Eng. 103, 108304 (2022)
Zhu, J.-Y., Park, T., Isola, P., Efros, A.A.: Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2223–2232 (2017)
Funding
This work was supported by the National funded postdoctoral researcher program (GZC20230403), the Harbin Science and Technology Bureau Manufacturing Innovation Talent Project (CXRC20221110393), the Heilongjiang Science and Technology Department Provincial Key R & D Program Applied Research Project (SC2022ZX06C0025), the Heilongjiang Science and Tech-nology Department Provincial Key R & D Program Guidance Project (GZ20220088).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Jing Zhao,Yongjun He, Zheng Zhi, Jian Qin, and Yining Xie. The first draft of the manuscript was written by Jing zhao and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., He, Yj., Shi, Z. et al. A style-aware network based on multi-task learning for multi-domain image normalization. Vis Comput (2024). https://doi.org/10.1007/s00371-024-03363-w
Accepted:
Published:
DOI: https://doi.org/10.1007/s00371-024-03363-w