Abstract
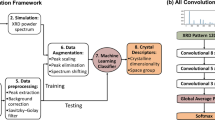
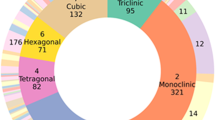
The experimental purpose of X-ray diffraction is to analyze crystalline material structure at the atomic and molecular levels. Such experiments are known as X-ray crystallography. Traditionally, human experts do it with some domain knowledge. X-ray crystallography is useful in numerous domains, e.g., physics, chemistry, and biology. It is tough to own manual physics of diffraction patterns to see a crystal structure with a colossal data set. A convolutional neural network (CNN) is a deep neural network that maps an input image into a high-dimensional space. CNN produces an affordable function for image classification. This paper uses an extension of the convolutional neural network to predict crystal structure from diffraction patterns. We propose a machine-enabled method to predict crystallographic size and space group from a limited number of XRD patterns for small films. We overcome the problem of scarce data within the development of building materials by combining the learning model of moderately monitored equipment, a physics information-enhancing strategy using data generated from the Inorganic Crystal Structure Database, and test data. We compare our approach with a large variety of typical addition as modern machine learning-based classification techniques for crystal structure prediction. Results show that our proposed system outperforms all the baselines by a significant margin for the crystal structure prediction task. Results also show the impact of the number of layers in the all-convolutional neural network architecture for crystal structure prediction.








Similar content being viewed by others
References
Agatonovic-Kustrin, S., Wu, V., Rades, T., Saville, D., Tucker, I.: Ranitidine hydrochloride x-ray assay using a neural network. J. Pharm. Biomed. Anal. 22(6), 985–992 (2000)
An, F.P., Liu, J.e., Bai, L.: Object recognition algorithm based on optimized nonlinear activation function-global convolutional neural network. The Visual Computer pp. 1–13 (2021)
Bouthillier, X., Konda, K., Vincent, P., Memisevic, R.: Dropout as data augmentation. arXiv preprint arXiv:1506.08700 (2015)
Gilmore, C.J., Barr, G., Paisley, J.: High-throughput powder diffraction i a new approach to qualitative and quantitative powder diffraction pattern analysis using full pattern profiles. J. Appl. Crystallogr. 37(2), 231–242 (2004)
Itano, W.M., Bollinger, J.J., Tan, J.N., Jelenković, B., Huang, X.P., Wineland, D.: Bragg diffraction from crystallized ion plasmas. Science 279(5351), 686–689 (1998)
Kim, Y.: Convolutional neural networks for sentence classification. arXiv preprint arXiv:1408.5882 (2014)
Kozuma, M., Deng, L., Hagley, E.W., Wen, J., Lutwak, R., Helmerson, K., Rolston, S., Phillips, W.D.: Coherent splitting of bose-einstein condensed atoms with optically induced bragg diffraction. Phys. Rev. Lett. 82(5), 871 (1999)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp. 1097–1105 (2012)
Lee, D., Lee, H., Jun, C.H., Chang, C.H.: A variable selection procedure for x-ray diffraction phase analysis. Appl. Spectrosc. 61(12), 1398–1403 (2007)
Obeidat, S.M., Al-Momani, I., Haddad, A., Bani Yasein, M.: Combination of icp-oes, xrf and xrd techniques for analysis of several dental ceramics and their identification using chemometrics. Spectroscopy 26(2), 141–149 (2011)
Park, J.K., Kang, D.J.: Unified convolutional neural network for direct facial keypoints detection. Visual Computer 35(11), 1615–1626 (2019)
Park, W.B., Chung, J., Jung, J., Sohn, K., Singh, S.P., Pyo, M., Shin, N., Sohn, K.S.: Classification of crystal structure using a convolutional neural network. IUCr J. 4(4), 486–494 (2017)
Press, W.H., Teukolsky, S.A.: Savitzky-golay smoothing filters. Computer Phys. 4(6), 669–672 (1990)
Seysen, M.: Simultaneous reduction of a lattice basis and its reciprocal basis. Combinatorica 13(3), 363–376 (1993)
Sheldrick, G.M.: Shelxt-integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A: Found. Adv. 71(1), 3–8 (2015)
Springenberg, J.T., Dosovitskiy, A., Brox, T., Riedmiller, M.A.: Striving for simplicity: The all convolutional net. CoRR abs/1412.6806 (2015)
Tatlier, M.: Artificial neural network methods for the prediction of framework crystal structures of zeolites from xrd data. Neural Comput. Appl. 20(3), 365–371 (2011)
Wang, J., Yang, Y., Mao, J., Huang, Z., Huang, C., Xu, W.: Cnn-rnn: A unified framework for multi-label image classification. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2285–2294 (2016)
Wei, Y., Xia, W., Lin, M., Huang, J., Ni, B., Dong, J., Zhao, Y., Yan, S.: Hcp: a flexible cnn framework for multi-label image classification. IEEE Trans. Pattern Anal. Mach. Intell. 38(9), 1901–1907 (2015)
Weidemüller, M., Hemmerich, A., Görlitz, A., Esslinger, T., Hänsch, T.W.: Bragg diffraction in an atomic lattice bound by light. Phys. Rev. Lett. 75(25), 4583 (1995)
Willis, B.: Lattice vibrations and the accurate determination of structure factors for the elastic scattering of x-rays and neutrons. Acta Crystallogr. Sect. A: Cryst. Phys., Diffr., Theor. General Crystallogr. 25(2), 277–300 (1969)
Zhou, X.Y., Zheng, J.Q., Li, P., Yang, G.Z.: Acnn: a full resolution dcnn for medical image segmentation. arXiv preprint arXiv:1901.09203 (2019)
Funding
No funding for the study involved.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The first author is an undergrad student and the second author is an Assistant Professor and advisor of the first author. Though we have financial support from the institute to register the paper in the journal, we have not received explicit funding from any company. We have conflict of interest with IITR domain (iitr.ac.in) and TCS domain (tcs.com) as the second author has worked at these places.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chakraborty, A., Sharma, R. A deep crystal structure identification system for X-ray diffraction patterns. Vis Comput 38, 1275–1282 (2022). https://doi.org/10.1007/s00371-021-02165-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-021-02165-8