Abstract
Purpose
Artificial intelligence is part of our daily life and machine learning techniques offer possibilities unknown until now in medicine. This study aims to offer an evaluation of the performance of machine learning (ML) techniques, for predicting bacterial resistance in a urology department.
Methods
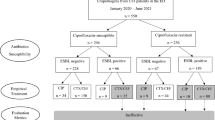
Data were retrieved from laboratory information system (LIS) concerning 239 patients with urolithiasis hospitalized in the urology department of a tertiary hospital over a 1-year period (2019): age, gender, Gram stain (positive, negative), bacterial species, sample type, antibiotics and antimicrobial susceptibility. In our experiments, we compared several classifiers following a tenfold cross-validation approach on 2 different versions of our dataset; the first contained only information of Gram stain, while the second had knowledge of bacterial species.
Results
The best results in the balanced dataset containing Gram stain, achieve a weighted average receiver operator curve (ROC) area of 0.768 and F-measure of 0.708, using a multinomial logistic regression model with a ridge estimator. The corresponding results of the balanced dataset, that contained bacterial species, achieve a weighted average ROC area of 0.874 and F-measure of 0.783, with a bagging classifier.
Conclusions
Artificial intelligence technology can be used for making predictions on antibiotic resistance patterns when knowing Gram staining with an accuracy of 77% and nearly 87% when identifying specific microorganisms. This knowledge can aid urologists prescribing the appropriate antibiotic 24–48 h before test results are known.
Similar content being viewed by others
Abbreviations
- ML:
-
Machine learning
- LIS:
-
Laboratory information system
- ROC:
-
Receiver operator curve
- UTI:
-
Urinary tract infections
- UCA-UTI:
-
Urinary calculi associated urinary tract infections
- AMR:
-
Multidrug antimicrobial resistance
- AI:
-
Artificial intelligence
- HIS:
-
Hospital information system
- CLSI:
-
Guidelines of laboratory standards institute
- MIC:
-
Minimum inhibitory concentration
- MMC:
-
Matthews correlation coefficient
- PRC:
-
Precision–recall plot
- AUC-PR:
-
Precision–recall curve
References
Holmgren K (1986) Urinary calculi and urinary tract infection. a clinical and microbiological study. Scand J Urol Nephrol Suppl 98:1–71
Chen D, Zhang Y, Huang J, Liang X, Zeng T, Lan C et al (2018) The analysis of microbial spectrum and antibiotic resistance of uropathogens isolated from patients with urinary stones. Int J Clin Pract 72(6):e13205
De Lorenzis E, Alba AB, Cepeda M, Galan JA, Geavlete P, Giannakopoulos S et al (2020) Bacterial spectrum and antibiotic resistance of urinary tract infections in patients treated for upper urinary tract calculi: a multicenter analysis. Eur J Clin Microbiol Infect Dis 39(10):1971–1981
Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP et al (2014) Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med 42(8):1749–1755
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S et al (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6):1589–1596
Yang B, Veneziano D, Somani BK (2020) Artificial intelligence in the diagnosis, treatment and prevention of urinary stones. Curr Opin Urol 30(6):782–787
Goodswen SJ, Barratt JLN, Kennedy PJ, Kaufer A, Calarco L, Ellis JT (2021) Machine learning and applications in microbiology. FEMS Microbiol Rev 45:15
Goh KH, Wang L, Yeow AYK, Poh H, Li K, Yeow JJL et al (2021) Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun 12(1):711
Wikler MA (2006) Performance standards for antimicrobial susceptibility testing. In: Sixteenth informational supplement M 100-S 16
World Medical Association Declaration of Helsinki (2013) ethical principles for medical research involving human subjects. JAMA 310(20):2191–2194
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software: an update. ACM SIGKDD Explor Newsl 11(1):10–18
Kasperczuk A, Dardzińska A (2016) Comparative evaluation of the different data mining techniques used for the medical database. Acta Mechanica et Automatica 10(3):233–238
Han J, Pei J, Yin Y (2000) Mining frequent patterns without candidate generation. ACM SIGMOD Rec 29(2):1–12
Tokas T, Herrmann TRW, Skolarikos A, Nagele U (2019) Pressure matters: intrarenal pressures during normal and pathological conditions, and impact of increased values to renal physiology. World J Urol 37(1):125–131
Tzelves L, Skolarikos A (2020) Suction use during endourological procedures. Curr Urol Rep 21(11):46
Feretzakis G, Sakagianni A, Loupelis E, Kalles D, Skarmoutsou N, Martsoukou M et al (2021) Machine learning for antibiotic resistance prediction: a prototype using off-the-shelf techniques and entry-level data to guide empiric antimicrobial therapy. Healthc Inform Res 27(3):214–221
Feretzakis G, Loupelis E, Sakagianni A, Kalles D, Lada M, Christopoulos C et al (2020) Using machine learning algorithms to predict antimicrobial resistance and assist empirical treatment. Stud Health Technol Inform 272:75–78
Feretzakis G, Loupelis E, Sakagianni A, Kalles D (2020) Using machine learning techniques to aid empirical antibiotic therapy decisions in the intensive care unit of a general hospital in greece. Antibiotics 9(2):50
Rajula HSR, Verlato G (2020) Comparison of conventional statistical methods with machine learning in medicine: diagnosis, drug development, and treatment. Medicina 56(9):455
Mitchell TM (1997) Machine learning. McGraw-Hill, New York
Yang Y, Niehaus KE, Walker TM, Iqbal Z, Walker AS, Wilson DJ et al (2018) Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 34(10):1666–1671
Kouchaki S, Yang Y, Walker TM, Sarah Walker A, Wilson DJ, Peto TEA et al (2019) Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinformatics 35(13):2276–2282
Farhat MR, Sultana R, Iartchouk O, Bozeman S, Galagan J, Sisk P et al (2016) Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am J Respir Crit Care Med 194(5):621–630
Längkvist M, Jendeberg J, Thunberg P, Loutfi A, Lidén M (2018) Computer aided detection of ureteral stones in thin slice computed tomography volumes using convolutional neural networks. Comput Biol Med 97:153–160
De Perrot T, Hofmeister J, Burgermeister S, Martin SP, Feutry G, Klein J et al (2019) Differentiating kidney stones from phleboliths in unenhanced low-dose computed tomography using radiomics and machine learning. Eur Radiol 29(9):4776–4782
Aminsharifi A, Irani D, Tayebi S, Kafash TJ, Shabanian T, Parsaei H (2020) Predicting the postoperative outcome of percutaneous nephrolithotomy with machine learning system: software validation and comparative analysis with guy’s stone score and the croes nomogram. J Endourol 34(6):692–699
Choo MS, Uhmn S, Kim JK, Han JH, Kim D-H, Kim J et al (2018) A prediction model using machine learning algorithm for assessing stone-free status after single session shock wave lithotripsy to treat ureteral stones. J Urol 200(6):1371–1377
Kazemi Y, Mirroshandel SA (2018) A novel method for predicting kidney stone type using ensemble learning. Artif Intell Med 84:117–126
Black KM, Law H, Aldoukhi A, Deng J, Ghani KR (2020) Deep learning computer vision algorithm for detecting kidney stone composition. BJUI Int 125(6):920–924
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization: LT, GF and IV, Methodology: GF, LL, Software EL, SB, Validation: DK, PM, Formal analysis: LL, LT, Investigation: MB, IM, IM, Resources: GF, Data curation: LL, MB, LT, Writing original draft preparation: GF, LT, LL, Writing-review and editing: IV, Visualization: DK, Supervision: AS, IV, Project administration: GF. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the institutional review board of Sismanogleio general hospital (Ref. No. 15178/2020).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tzelves, L., Lazarou, L., Feretzakis, G. et al. Using machine learning techniques to predict antimicrobial resistance in stone disease patients. World J Urol 40, 1731–1736 (2022). https://doi.org/10.1007/s00345-022-04043-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-022-04043-x