Abstract
The GARP transcription factors have been identified for multiple biological functions throughout the life cycle of a plant. Despite of its involvement in crucial functions, systematic study of GARPs remains obscure in plants. In this study, we explored the genomic, molecular, and evolutionary perspectives of the GARP gene family in the three major leguminous plants, namely chickpea (Cicer arietinum), soybean (Glycine max), and barrel clover (Medicago truncatula). Here, we identified 53, 56, and 107 GARP genes in Cicer, Medicago, and Glycine, respectively. They were classified into four clades and two sub-clades as per phylogenetic analysis, and the result was supported by consensus motifs, domain organization, and exon–intron structures. Detailed comparative analysis indicates conservation of the GARP gene family in plants. Identification of paralogous and orthologous gene pairs revealed that the expansion of this family occurs mainly through genome duplication in legumes. Additionally, the three-dimensional structure and functional enrichment analysis indicated their major role in signaling, growth, development, and stress processes. The chickpea GARP genes were also characterized for their transcript modulation in diverse plant organs and during pathogenic stress. Differential regulation of 24 CaGARP genes was observed during Ascochyta Blight (AB) stress. Characterization of AB-responsive genes reveals an over-representation of stress and hormone-binding elements on the promoter of CaGARPs. Additionally, interactome analysis also confirms the role of GARPs in plant stress and development. Our findings not only provide a handful of stress-responsive genes but also lay the foundation for prospective functional studies of GARPs in legumes.








Similar content being viewed by others
Data Availability
All relevant data can be found within the manuscript and its supplementary materials.
References
Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20(8):2102–2116
Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10(10):771
Chang WC, Lee TY, Huang HD, Huang HY, Pan RL (2008) PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9(1):1–14
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN (2011) Brother of Lux Arrhythmo is a component of the Arabidopsis circadian clock. Plant Cell 23(3):961–972
Davidson EH, Jacobs HT, Britten RJ (1983) Eukaryotic gene expression: very short repeats and coordinate induction of genes. Nature 301(5900):468–470
Hai Du, Wang YB, Xie YI, Zhe L, Jiang SJ, Zhang SS, Huang YB, Tang YX (2013) Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res 20(5):437–448
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13(20):1768–1774
Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11(16):1251–1260
Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31(6):713–727
Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND (2005) Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference 1228–1235
Hall LN, Rossini L, Cribb L, Langdale JA (1998) GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10(6):925–936
Han XY, Li PX, Zou LJ, Tan WR, Zheng T, Zhang DW, Lin HH (2016) GOLDEN2-LIKE transcription factors coordinate the tolerance to Cucumber mosaic virus in Arabidopsis. Biochem Bioph Res Co 477(4):626–632
Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant MYB-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14(9):2015–2029
Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera-Rauh F, Hokin SA, Barton MK, Kerstetter RA (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26(1):246–262
Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45(D1):D1040–D1045
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411(6838):706–709
Kesarwani M, Yoo J, Dong X (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144(1):336–346
Kiba T, Inaba J, Kudo T, Ueda N, Konishi M, Mitsuda N, Takiguchi Y, Kondou Y, Yoshizumi T, Ohme-Takagi M, Matsui M (2018) Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell 30(4):925–945
Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li C, Ng CKY, Fan LM (2015) MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot 114:80–91
Liu H, Yang H, Wu C, Feng J, Liu X, Qin H, Wang D (2009) Overexpressing HRS1 confers hypersensitivity to low phosphate-elicited inhibition of primary root growth in Arabidopsis thaliana. J Integr Plant Biol 51(4):382–392
Liu Y, Zeng Y, Li Y, Liu Z, Lin-Wang K, Espley RV, Allan AC, Zhang J (2020) Genomic survey and gene expression analysis of the MYB-related transcription factor superfamily in potato (Solanum tuberosum L.). Int J Biol Macromol 164:2450–2464
Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41(6):791–803
Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G (2015) AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun 6(1):1–11
Murmu J, Wilton M, Allard G, Pandeya R, Desveaux D, Singh J, Subramaniam R (2014) Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol Plant Pathol 15(2):174–184
Nagarajan VK, Satheesh V, Poling MD, Raghothama KG, Jain A (2016) Arabidopsis MYB-related HHO2 exerts a regulatory influence on a subset of root traits and genes governing phosphate homeostasis. Plant Cell Physiol 57(6):1142–1152
Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10(10):963–972
Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, Lopez-Baltazar J (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336(6089):1711–1715
Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, Leyva A (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. P Natl Acad Sci USA 111(41):14947–14952
Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290(5499):2105–2110
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Gene Dev 15(16):2122–2133
Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B, Krouk G (2017) The world according to GARP transcription factors. Curr Opin Plant Biol 39:159–167
Savitch LV, Subramaniam R, Allard GC, Singh J (2007) The GLK1 ‘regulon’encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochem Bioph Res Co 359(2):234–238
Sawaki N, Tsujimoto R, Shigyo M, Konishi M, Toki S, Fujiwara T, Yanagisawa S (2013) A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol 54(4):506–517
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D (2010) Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385
Sharma M, Ghosh R (2016) An update on genetic resistance of chickpea to Ascochyta blight. Agronomy 6(1):18
Singh R, Dwivedi A, Singh Y, Kumar K, Ranjan A, Verma PK (2022) Transcriptome analysis reveals robust transcriptional reprogramming during early stages of Cicer-Ascochyta interaction. bioRxiv. https://doi.org/10.1101/2022.03.26.485904
Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170(1):499–514
Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45(1):28–39
Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, Mayer KF (2014) An improved genome release (version Mt4. 0) for the model legume Medicago truncatula. BMC Genomics 15(1):1–14
Tian F, Yang DC, Meng YQ, Jin J, Gao G (2020) PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res 48(D1):D1104–D1113
Valliyodan B, Cannon SB, Bayer PE, Shu S, Brown AV, Ren L, Jenkins J, Chung CY, Chan TF, Daum CG, Plott C et al (2019) Construction and comparison of three reference-quality genome assemblies for soybean. Plant J 100(5):1066–1082
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, Millan T (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotechnol 31(3):240–246
Verma M, Kumar V, Patel RK, Garg R, Jain M (2015) CTDB: an integrated chickpea transcriptome database for functional and applied genomics. PLoS ONE 10(8):e0136880
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21(4):1109–1128
William Roy S, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7(3):211–221
Wu C, Feng J, Wang R, Liu H, Yang H, Rodriguez PL, Qin H, Liu X, Wang D (2012) HRS1 acts as a negative regulator of abscisic acid signaling to promote timely germination of Arabidopsis seeds. PLoS ONE 7(4):e35764
Wu G, Liu S, Zhao Y, Wang W, Kong Z, Tang D (2015) ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27(3):857–873
Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. P Natl Acad Sci USA 96(26):15336–15341
Yan S, Yan CJ, Zeng XH, Yang YC, Fang YW, Tian CY, Sun YW, Cheng ZK, Gu MH (2008) ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol Biol 68(3):239–250
Yanagisawa S (2013) Characterization of a nitrate-inducible transcriptional repressor NIGT1 provides new insights into DNA recognition by the GARP family proteins. Plant Signal Behav 8(6):506–517
Yang X, Guo T, Li J, Chen Z, Guo B, An X (2021) Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int J Biol Macromol 191:359–376
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60(1):107–124
Yokoyama A, Yamashino T, Amano YI, Tajima Y, Imamura A, Sakakibara H, Mizuno T (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48(1):84–96
Acknowledgements
RS acknowledges University Grants Commission (UGC), India for the SRF fellowship. The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Contributions
Conception and design—RS and PKV, analysis and interpretation of the data—RS, drafting of the article—RS, critical revision of the article for important intellectual content—AP and PKV, final approval of the article—PKV, obtaining of funding—PKV.
Corresponding author
Additional information
Handling Editor: Padmanabh Dwivedi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2022_10746_MOESM1_ESM.tiff
Supplementary file1 Fig. S1. Multiple sequence alignment of GARP B-motif. (A) Cicer, (B) Medicago, and (C) Glycine. Amino acid sequences of GARP B-motif were aligned using the ClustalX2.1 (http://www.clustal.org) program and visualized through Jalview software (Waterhouse et al., 2009). The conserved GARP motif is boxed in blue color and the highlighted colored region indicates the conserved amino acid residues among the sequences (TIFF 15342 kb)
344_2022_10746_MOESM2_ESM.tiff
Supplementary file2 Fig. S2. The logo and sequences of the conserved motifs are identified in three legumes. (A) Cicer, (B) Medicago, and (C) Glycine. The motifs are predicted using MEME v5.4.1 (TIFF 16487 kb)
344_2022_10746_MOESM3_ESM.tiff
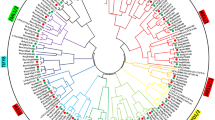
Supplementary file3 Fig. S3. Prediction of stress-responsive GARP genes. The phylogenetic tree of identified GARP genes is prepared by utilizing functionally characterized GARP members. Representation of the phylogenetic tree of (A) Cicer, (B) Medicago, and (C) Glycine. The tree was constructed using the neighbor-joining method in Mega7 (Kumar, Stecher, and Tamura, 2016). Bootstrap values are obtained from 1000 iterations and indicated by circles on each branch. The branches of different clades are marked in different colors (TIFF 11979 kb)
344_2022_10746_MOESM4_ESM.tiff
Supplementary file4 Fig. S4. Predicted promoter element for putative AB-stress-responsive CaGARPs. The 1500 bp promoter sequences are analyzed through PlantPan (Chang et al., 2008) and PlantCare databases (Lescot et al., 2002) database. The scale represents the position of promoter elements. The lines denote promoter sequences and identified elements are represented by different color boxes (TIFF 12868 kb)
344_2022_10746_MOESM5_ESM.tiff
Supplementary file5 Fig. S5. Representation of protein-protein interaction network. CaGARPs are mapped on Arabidopsis STRINGDB (https://string-db.org). The network is visualized through Cytoscape v3.6.2 (TIFF 4583 kb)
344_2022_10746_MOESM9_ESM.docx
Supplementary file9 Table S4. The number of identified GARPs, the total number of annotated genes, and genome size of Cicer, Medicago, and Glycine (DOCX 13 kb)
344_2022_10746_MOESM12_ESM.docx
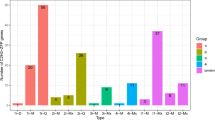
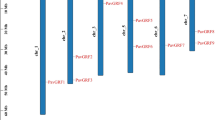
Supplementary file12 Table S7. Fold change of the differentially expressed GARP genes during AB-stress in chickpea (DOCX 19 kb)
344_2022_10746_MOESM13_ESM.xlsx
Supplementary file13 Table S8. Identified promoter elements for putative AB-stress-responsive CaGARP genes (XLSX 140 kb)
344_2022_10746_MOESM14_ESM.xlsx
Supplementary file14 Table S9. Predicted transcription factor binding sites for putative AB-stress-responsive CaGARP genes (XLSX 348 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, R., Pandey, A. & Verma, P.K. Comparative Genomic Analysis of GARP Transcription Factor Family in Legumes and Identification of Stress-Responsive Candidate Genes. J Plant Growth Regul 42, 6005–6020 (2023). https://doi.org/10.1007/s00344-022-10746-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-022-10746-7