Abstract
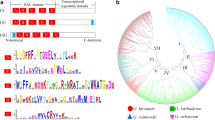
GT factors, also called trihelix transcription factors (TFs), belong to a small family of TFs and were first discovered because of their unique binding to GT elements. Recent studies have demonstrated the functions of GT factors in plant development and stress. Cotton is a major source of fiber and oilseed. However, studies about the GT factors in cotton are limited, especially with regard to their function and evolution. Therefore, it is important to study the GT factors in cotton. This study was based on three genome databases of cotton (A, D, and AD) and the GT factors in Arabidopsis. We retrieved 39 putative GhGT genes in Gossypium hirsutum, 14 GaGT in G. arboreum, and 14 GrGT in G. raimondii. For 39 GhGT genes, we characterized their motif features, domain sequence alignments, and phylogenetic relationship with other species. To understand the functions of GT factors in the senescent cotton leaves, we isolated eight GhGT genes from yellow leaves. In addition, we detected the expression patterns of these eight genes in that particular tissue as well as their effects on fiber development and responses to stresses and plant hormones, especially in leaf development, using real-time quantitative PCR. In addition, we also studied the function of GhGT31 in Arabidopsis. The results not only laid the foundation for an understanding of the functions of the GT factors in cotton but also provided significant information about the effects of GT factors on cotton development and on the responses to various stresses.
Graphical Abstract











Similar content being viewed by others
References
Agarwal PK, Shukla PS, Gupta K, Jha B (2013) Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol 54:102–123. doi:10.1007/s12033-012-9538-3
Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056. doi:10.1104/pp.123.3.1047
Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65:3993–4008. doi:10.1093/jxb/eru173
Amagai A, Maeda Y (1992) The ethylene action in the development of cellular slime molds: an analogy to higher plants. Protoplasma 167:159–168. doi:10.1007/BF01403379
Ayadi M, Delaporte V, Li YF, Zhou DX (2004) Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett 562:147–154. doi:10.1016/S0014-5793(04)00222-4
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264. doi:10.1111/j.1365-313X.2010.04151.x
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Breuer C, Kawamura A, Ichikawa T, Tominaga-Wada R, Wada T, Kondou Y, Muto S, Matsui M, Sugimoto K (2009) The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21:2307–2322. doi:10.1105/tpc.109.068387
Brewer PB, Howles PA, Dorian K, Griffith ME, Ishida T, Kaplan-Levy RN, Kilinc A, Smyth DR (2004) Petal loss, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131:4035–4045. doi:10.1242/dev.01279
Caro E, Desvoyes B, Gutierrez C (2012) GTL1 keeps cell growth and nuclear ploidy under control. EMBO J 31:4483–4485. doi:10.1038/emboj.2012.311
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape J-, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145:1303–1310. doi:10.1104/pp.107.107672
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi:10.1046/j.1365-313x.1998.00343.x
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679. doi:10.1146/annurev-arplant-042809-112122
Degani O, Drori R, Goldblat Y (2015) Plant growth hormones suppress the development of Harpophora maydis, the cause of late wilt in maize. Physiol Mol Biol Plants 21:137–149. doi:10.1007/s12298-014-0265-z
Dehesh K, Bruce WB, Quail PH (1990) A trans-acting factor that binds to a GT-motif in a phytochrome gene promoter. Science 250:1397–1399. doi:10.1126/science.2255908
Dehesh K, Hung H, Tepperman JM, Quail PH (1992) GT-2: a transcription factor with twin autonomous DNA-binding domains of closely related but different target sequence specificity. EMBO J 11:4131–4144
Dhandapani G, Lakshmi Prabha A, Kanakachari M, Phanindra ML, Prabhakaran N, Gothandapani S, Padmalatha KV, Solanke AU, Kumar PA (2015) GhDRIN1, a novel drought-induced gene of upland cotton (Gossypium hirsutum L.) confers abiotic and biotic stress tolerance in transgenic tobacco. Biotechnol Lett 37:907–919. doi:10.1007/s10529-014-1733-9
Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ (2014) Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J 79:13–27. doi:10.1111/tpj.12538
Du H, Wang YB, Xie Y, Liang Z, Jiang SJ, Zhang SS, Huang YB, Tang YX (2013) Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res 20:437–448. doi:10.1093/dnares/dst021
Duan Q, Jiang W, Ding M, Lin Y, Huang D (2014) Light affects the chloroplast ultrastructure and post-storage photosynthetic performance of watermelon (Citrullus lanatus) plug seedlings. PLoS One 9:e111165. doi:10.1371/journal.pone.0111165
Fang Y, Xie K, Hou X, Hu H, Xiong L (2010) Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol Genet Genomics 283:157–169. doi:10.1007/s00438-009-0507-x
Fatma M, Khan NA (2014) The inhibitory effect of salinity on the photosynthetic machinery of mustard plant and the alleviation of this effect by ethylene. Toxicol Lett 229:S124–S124. doi:10.1016/j.toxlet.2014.06.443
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319. doi:10.1093/jxb/erh003
Gao MJ, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, Rozwadowski K (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21:54–71. doi:10.1105/tpc.108.061309
Geraldo N, Bäurle I, Kidou S, Hu X, Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150:1611–1618. doi:10.1104/pp.109.137448
Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua N-H (1992) Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 4:839–849. doi:10.1105/tpc.4.7.839
Giuntoli B, Lee SC, Licausi F, Kosmacz M, Oosumi T, van Dongen JT, Bailey-Serres J, Perata P (2014) A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol 12:e1001950. doi:10.1371/journal.pbio.1001950
Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–818. doi:10.1126/science.1185383
Green PJ, Kay SA, Chua NH (1987) Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6:2543–2549
Groen SC, Whiteman NK (2014) The evolution of ethylene signaling in plant chemical ecology. J Chem Ecol 40:700–716. doi:10.1007/s10886-014-0474-5
Hajouj T, Michelis R, Gepstein S (2000) Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol 124:1305–1314. doi:10.1104/pp.124.3.1305
Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53:927–937. doi:10.1093/jexbot/53.370.927
Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139:267–274. doi:10.1104/pp.105.065698
Kaplan-Levy RN, Brewer PB, Quon T, Smyth DR (2012) The trihelix family of transcription factors—light, stress and development. Trends Plant Sci 17:163–171. doi:10.1016/j.tplants.2011.12.002
Kaur C, Kushwaha HR, Mustafiz A, Pareek A, Sopory SK, Singla-Pareek SL (2015) Analysis of global gene expression profile of rice in response to methylglyoxal indicates its possible role as a stress signal molecule. Front Plant Sci 6:682. doi:10.3389/fpls.2015.00682
Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, Narasimhan ML (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62:3981–3992. doi:10.1093/jxb/err094
Kramer GF, Norman HA, Krizek DT, Mirecki RM (1991) Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 30:2101–2108. doi:10.1016/0031-9422(91)83595-C
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) CIRCOS: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi:10.1101/gr.092759.109
Kuhn RM, Caspar T, Dehesh K, Quail PH (1993) DNA binding factor GT-2 from Arabidopsis. Plant Mol Biol 23:337–348. doi:10.1007/BF00029009
Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, Okada K, Shinozaki K (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47:640–651. doi:10.1111/j.1365-313X.2006.02808.x
LeGourrierec J, Li YF, Zhou DX (1999) Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA–TBP–TATA complex. Plant J 18:663–668. doi:10.1046/j.1365-313x.1999.00482.x
Li X, Qin G, Chen Z, Gu H, Qu LJ (2008) A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol Biol 66:315–327. doi:10.1007/s11103-007-9272-6
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572. doi:10.1038/ng.2987
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015) Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530. doi:10.1038/nbt.3208
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. doi:10.1146/annurev.arplant.57.032905.105316
Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226:11–20. doi:10.1007/s00425-006-0460-4
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem 262:247–257. doi:10.1046/j.1432-1327.1999.00349.x
Liu B, Zhu Y, Zhang T (2015) The R3-MYB gene GhCPC negatively regulates cotton fiber elongation. PLoS One 10:e0116272. doi:10.1371/journal.pone.0116272
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Long L, Gao W, Xu L, Liu M, Luo X, He X, Yang X, Zhang X, Zhu L (2014) GbMPK3, a mitogen-activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco. Plant Cell Tissue Organ Cult 116:153–162. doi:10.1007/s11240-013-0392-1
Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y (2014) Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant 7:1670–1682. doi:10.1093/mp/ssu085
Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59:52–62. doi:10.1111/j.1365-313X.2009.03847.x
Mulkey TJ, Evans ML, Kuzmanoff KM (1983) The kinetics of abscisic acid action on root growth and gravitropism. Planta 157:150–157. doi:10.1007/BF00393649
Nagano Y (2000) Several features of the GT-factor trihelix domain resemble those of the Myb DNA-binding domain. Plant Physiol 124:491–493. doi:10.1104/pp.124.2.491
Nagano Y, Yamamoto Y, Inaba T, Sasaki Y (2002) Functional characterization of a DNA-binding protein DF1. Plant Cell Physiol 43:S228–S228
Nagata T, Niyada E, Fujimoto N, Nagasaki Y, Noto K, Miyanoiri Y, Murata J, Hiratsuka K, Katahira M (2010) Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins 78:3033–3047. doi:10.1002/prot.22827
Nakashima K, Yamaguchi-Shinozaki K (2006) Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant 126:62–71. doi:10.1111/j.1399-3054.2005.00592.x
Ni M, Dehesh K, Tepperman JM, Quail PH (1996) GT-2: in vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8:1041–1059. doi:10.1105/tpc.8.6.1041
O’Grady K, Goekjian VH, Naim CJ, Nagao RT, Key JL (2001) The transcript abundance of GmGT-2, a new member of the GT-2 family of transcription factors from soybean, is down-regulated by light in a phytochrome-dependent manner. Plant Mol Biol 47:367–378. doi:10.1023/A:1011629307051
Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132:603–614
Parida AK, Das AB (2005) Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf 60:324–349. doi:10.1016/j.ecoenv.2004.06.010
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee SH, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135:2150–2161. doi:10.1104/pp.104.041442
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li JP, Lin LF, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang HB, Xu CM, Wang JP, Wang ZN, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, Rahman MU, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MFS, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang TZ, Dennis ES, Mayer KFX, Peterson DG, Rokhsar DS, Wang XY, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427. doi:10.1038/nature11798
Peng X, Zhao Y, Cao J, Zhang W, Jiang H, Li X, Ma Q, Zhu S, Cheng B (2012) CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One 7:e40120. doi:10.1371/journal.pone.0040120
Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B (2009) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38(Suppl 1):D822–D827. doi:10.1093/nar/gkp805
Perisic O, Lam E (1992) A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 4:831–838. doi:10.1105/tpc.4.7.831
Porcel R, Zamarreño ÁM, García-Mina JM, Aroca R (2014) Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol 14:36. doi:10.1186/1471-2229-14-36
Posada D (2009) Bioinformatics for DNA sequence analysis. In: Methods in molecular biology, vol 537. Humana Press, New York
Pu L, Li Q, Fan X, Yang W, Xue Y (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180:811–820. doi:10.1534/genetics.108.093070
Qin Y, Ma X, Yu G, Wang Q, Wang L, Kong L, Kim W, Wang HW (2014) Evolutionary history of trihelix family and their functional diversification. DNA Res 21:499–510. doi:10.1093/dnares/dsu016
Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5:278–282. doi:10.1016/S1360-1385(00)01655-1
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401. doi:10.1016/j.tplants.2010.04.006
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110. doi:10.1126/science.290.5499.2105
Schweiger R, Heise AM, Persicke M, Müller C (2014) Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant Cell Environ 37:1574–1585. doi:10.1111/pce.12257
Shangguan XX, Xu B, Yu ZX, Wang LJ, Chen XY (2008) Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. J Exp Bot 59:3533–3542. doi:10.1093/jxb/ern204
Shinohara T, Leskovar DI (2014) Effects of ABA, antitranspirants, heat and drought stress on plant growth, physiology and water status of artichoke transplants. Sci Hortic 165:225–234. doi:10.1016/j.scienta.2013.10.045
Singh B, Avci U, Eichler Inwood SE, Grimson MJ, Landgraf J, Mohnen D, Sørensen I, Wilkerson CG, Willats WG, Haigler CH (2009) A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol 150:684–699. doi:10.1104/pp.109.135459
Song M, Fan S, Pang C, Wei H, Yu S (2014) Genetic analysis of the antioxidant enzymes, methane dicarboxylic aldehyde (MDA) and chlorophyll content in leaves of the short season cotton (Gossypium hirsutum L.). Euphytica 198:153–162. doi:10.1007/s10681-014-1100-x
Suo J, Liang X, Pu L, Zhang Y, Xue Y (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta 1630:25–34. doi:10.1016/j.bbaexp.2003.08.009
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Tu L, Zhang X, Liu D, Jin S, Cao J, Zhu L, Deng F, Tan J, Zhang C (2007) Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin Sci Bull 52:3110–3117. doi:10.1007/s11434-007-0461-0
Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135:1206–1220. doi:10.1104/pp.104.045179
Wang WX, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14. doi:10.1105/tpc.104.024844
Wang R, Hong G, Han B (2004a) Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 324:105–115. doi:10.1016/j.gene.2003.09.008
Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY (2004b) Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16:2323–2334
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1103. doi:10.1038/ng.2371
Wang XH, Li QT, Chen HW, Zhang WK, Ma B, Chen SY, Zhang JS (2014) Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol 14:339. doi:10.1186/s12870-014-0339-7
Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697. doi:10.1093/aob/mcm079
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in annals of botany. Ann Bot 111:1021–1058. doi:10.1093/aob/mct067
Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13:495–502. doi:10.1016/j.pbi.2010.09.007
Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis della RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24:3307–3319. doi:10.1105/tpc.112.101428
Willmann MR, Mehalick AJ, Packer RL, Jenik PD (2011) MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol 155:1871–1884. doi:10.1104/pp.110.171355
Wu R, Li S, He S, Wassmann F, Yu C, Qin G, Schreiber L, Qu LJ, Gu H (2011) CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23:3392–3411. doi:10.1105/tpc.111.088625
Xie ZM, Zou HF, Lei G, Wei W, Zhou QY, Niu CF, Liao Y, Tian AG, Ma B, Zhang WK, Zhang JS, Chen SY (2009) Soybean trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS One 4:e6898. doi:10.1371/journal.pone.0006898
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183
Xu Z, Yu JZ, Cho J, Yu J, Kohel RJ, Percy RG (2010) Polyploidization altered gene functions in cotton (Gossypium spp.). PLoS One 5:e14351. doi:10.1371/journal.pone.0014351
Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV (2010) The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22:4128–4141. doi:10.1105/tpc.110.078691
Yu J, Jung S, Cheng CH, Ficklin SP, Lee T, Zheng P, Jones D, Percy RG, Main D (2014) CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res 42:D1229–D1236. doi:10.1093/nar/gkt1064
Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39:D1114–D1117. doi:10.1093/nar/gkq1141
Zhang J, Zhu L, Yu S, Jin Q (2014) Involvement of 1-methylcyclopropene in plant growth, ethylene production, and synthase activity of inferior spikelets in hybrid rice differing in panicle architectures. J Plant Growth Regul 33:551–561. doi:10.1007/s00344-013-9404-y
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537. doi:10.1038/nbt.3207
Zhao J, Li S, Jiang T, Liu Z, Zhang W, Jian G, Qi F (2012) Chilling stress—the key predisposing factor for causing Alternaria alternata infection and leading to cotton (Gossypium hirsutum L.) leaf senescence. PLoS One 7:e36126. doi:10.1371/journal.pone.0036126
Zhou DX (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4:210–214. doi:10.1016/S1360-1385(99)01418-1
Acknowledgments
We thank The National High Technology Research and Development Program of China (Grant No. 2013AA102601) for the financial support provided to this project.
Author's contributions
SXY, SLF, CYP, and MZS designed the experiments. HLW collected all the sequences, YNG performed the experiments and wrote the manuscript, and LLD helped in analyzing the data. CYP and OE revised the language. All the authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Conserved domains multiple alignments of 39 GhGT factors through DNAMAN software
Supplementary Fig. 2 MDA content of cotyledons of CCRI10 and Liao4086 during leaf senescence
Supplementary Fig. 3 Chlorophyll content of cotyledon and true leaf during aging
Supplementary Fig. 4 Soluble protein content of CCRI10 and Liao4086 during leaf senescence
Rights and permissions
About this article
Cite this article
Guo, Y., Dou, L., Evans, O. et al. Identification of GT Factors in Response to Stresses and Leaf Senescence in Gossypium hirsutum L.. J Plant Growth Regul 36, 22–42 (2017). https://doi.org/10.1007/s00344-016-9619-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-016-9619-9