Abstract.
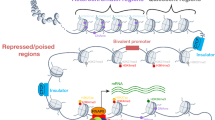
Gene expression is presently a major focus in genome analysis, and the experimental data on regulatory mechanisms and functional transcription factor binding sites are steadily growing. However, the annotation of transcriptional regulation of sequences cannot keep pace with the exponential growth of sequence databases. Employing detailed experimental data of a single promoter or enhancer to predict genes with similar regulation would provide a powerful method to link the literature about transcriptional regulation and sequence databases. To this end, we used information on individual functional transcription factor binding sites to compose in silico promoter and enhancer models of muscle-specific genes and to analyze the rodents section of EMBL with these models. Exhaustive evaluation of all hits revealed every second to third match to be a muscle-associated gene. Moreover, functionally related regulatory regions were detected by our model-based approach even in the absence of sequence similarity. We believe that this new approach is a substanial extension to database analysis by BLAST or FASTA, which are restricted to sequence similarity.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 18 April 2000 / Accepted: 17 August 2000
Rights and permissions
About this article
Cite this article
Gailus-Durner, V., Scherf, M. & Werner, T. Experimental data of a single promoter can be used for in silico detection of genes with related regulation in the absence of sequence similarity. 12, 67–72 (2001). https://doi.org/10.1007/s003350010219
Issue Date:
DOI: https://doi.org/10.1007/s003350010219