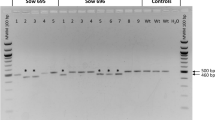
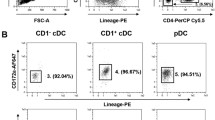
Abstract
Studies in mice genetically lacking natural killer T (NKT) cells show that these lymphocytes make important contributions to both innate and adaptive immune responses. However, the usefulness of murine models to study human NKT cells is limited by the many differences between mice and humans, including that their NKT cell frequencies, subsets, and distribution are dissimilar. A more suitable model may be swine that share many metabolic, physiological, and growth characteristics with humans and are also similar for NKT cells. Thus, we analyzed genetically modified pigs made deficient for CD1d that is required for the development of Type I invariant NKT (iNKT) cells that express a semi-invariant T-cell receptor (TCR) and Type II NKT cells that use variable TCRs. Peripheral blood analyzed by flow cytometry and interferon-γ enzyme-linked immuno spot assays demonstrated that CD1d-knockout pigs completely lack iNKT cells, while other leukocyte populations remain intact. CD1d and NKT cells have been shown to be involved in shaping the composition of the commensal microbiota in mice. Therefore, we also compared the fecal microbiota profile between pigs expressing and lacking NKT cells. However, no differences were found between pigs lacking or expressing CD1d. Our results are the first to show that knocking-out CD1d prevents the development of NKT cells in a non-rodent species. CD1d-deficient pigs should offer a useful model to more accurately determine the contribution of NKT cells for human immune responses. They also have potential for understanding how NKT cells impact the health of commercial swine.


Similar content being viewed by others
References
Artiaga BL, Whitener RL, Staples CR, Driver JP (2014) Adjuvant effects of therapeutic glycolipids administered to a cohort of NKT cell-diverse pigs. Vet Immunol Immunopathol 162:1–13
Bendelac A, Savage PB, Teyton L (2007) The biology of NKT cells. Annu Rev Immunol 25:297–336
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M (1997) Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science 278:1623–1626
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Eguchi-Ogawa T, Morozumi T, Tanaka M, Shinkai H, Okumura N, Suzuki K, Awata T, Uenishi H (2007) Analysis of the genomic structure of the porcine CD1 gene cluster. Genomics 89:248–261
Godfrey DI, Stankovic S, Baxter AG (2010) Raising the NKT cell family. Nat Immunol 11:197–206
Kinjo Y, Kitano N, Kronenberg M (2013) The role of invariant natural killer T cells in microbial immunity. J Infect Chemother 19:560–570
Kumar V, Delovitch TL (2014) Different subsets of natural killer T cells may vary in their roles in health and disease. Immunology 142:321–336
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L (1997) CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity 6:469–477
Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, Koga Y, Samsom JN, Oshima K, Kikuchi M, Escher JC, Hattori M, Onderdonk AB, Blumberg RS (2009) Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Investig 119:1241–1250
Park SH, Bendelac A (2000) CD1-restricted T-cell responses and microbial infection. Nature 406:788–792
Pillai AB, George TI, Dutt S, Teo P, Strober S (2007) Host NKT cells can prevent graft-versus-host disease and permit graft antitumor activity after bone marrow transplantation. J Immunol 178:6242–6251
Robertson FC, Berzofsky JA, Terabe M (2014) NKT cell networks in the regulation of tumor immunity. Front Immunol 5:543
Thierry A, Robin A, Giraud S, Minouflet S, Barra A, Bridoux F, Hauet T, Touchard G, Herbelin A, Gombert JM (2012) Identification of invariant natural killer T cells in porcine peripheral blood. Vet Immunol Immunopathol 149:272–279
Van Kaer L, Parekh VV, Wu L (2011) Invariant natural killer T cells: bridging innate and adaptive immunity. Cell Tissue Res 343:43–55
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, Anderson G (2014) An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol 192:2659–2666
Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O’Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS (2014) Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod 91:78
Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812
Acknowledgments
This work was supported by the National Swine Resource and Research Center, UF Research Opportunity Seed Fund, and IFAS Early Career Seed Fund awards. The National Institutes of Health Tetramer Core Facility provided the CD1d tetramers. G.Y. is supported by a UF Animal Molecular and Cellular Biology graduate fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guan Yang and Bianca L. Artiaga contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, G., Artiaga, B.L., Hackmann, T.J. et al. Targeted disruption of CD1d prevents NKT cell development in pigs. Mamm Genome 26, 264–270 (2015). https://doi.org/10.1007/s00335-015-9564-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-015-9564-0